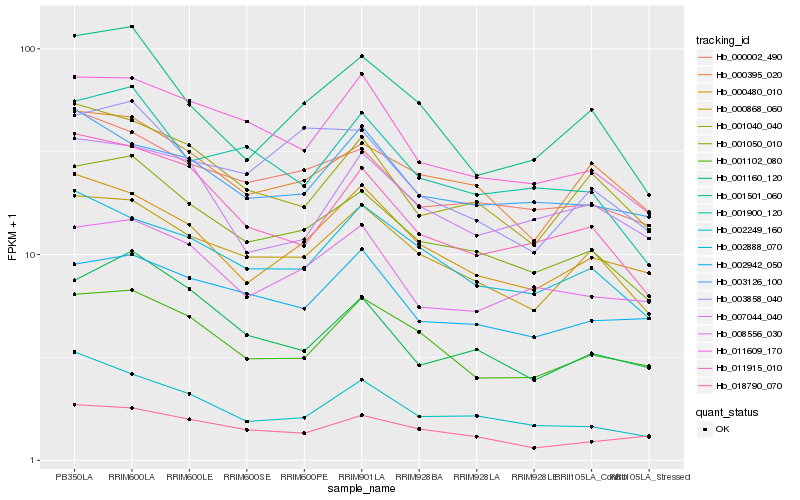
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001050_010 |
0.0 |
- |
- |
Advillin [Gossypium arboreum] |
| 2 |
Hb_000868_060 |
0.0650220395 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 3 |
Hb_000002_490 |
0.0730073272 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 4 |
Hb_011915_010 |
0.0747024395 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 5 |
Hb_001900_120 |
0.0750106186 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 6 |
Hb_011609_170 |
0.0775114929 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 7 |
Hb_000395_020 |
0.0789647085 |
- |
- |
PREDICTED: ankyrin repeat and KH domain-containing protein R11A8.7 [Jatropha curcas] |
| 8 |
Hb_001040_040 |
0.0803791007 |
- |
- |
Uncharacterized protein TCM_029905 [Theobroma cacao] |
| 9 |
Hb_001102_080 |
0.0821693561 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 4 [Jatropha curcas] |
| 10 |
Hb_001160_120 |
0.0833714278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002888_070 |
0.0835589223 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 12 |
Hb_001501_060 |
0.0857105686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000480_010 |
0.0880656799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002249_160 |
0.0883146295 |
- |
- |
- |
| 15 |
Hb_018790_070 |
0.0885161947 |
- |
- |
PREDICTED: uncharacterized protein LOC105650163 [Jatropha curcas] |
| 16 |
Hb_008556_030 |
0.0886554319 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003858_040 |
0.0923183901 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 18 |
Hb_003126_100 |
0.0957846613 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 19 |
Hb_002942_050 |
0.0996863691 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 20 |
Hb_007044_040 |
0.1013405695 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |