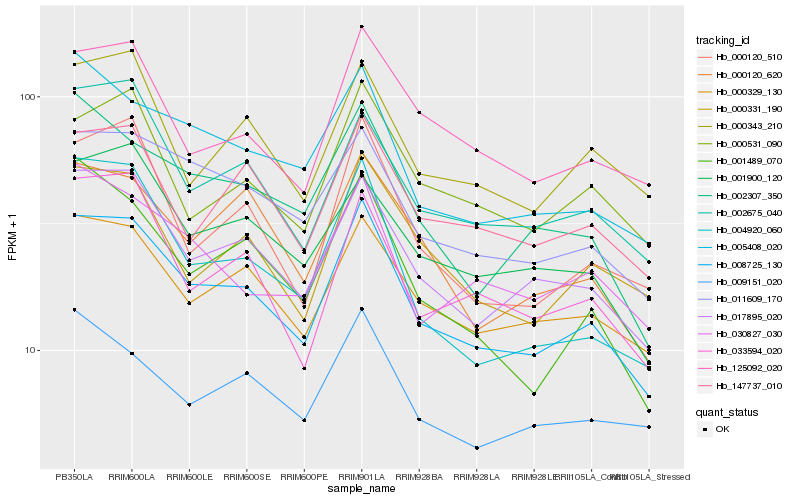
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008725_130 |
0.0 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 2 |
Hb_017895_020 |
0.0567583674 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 3 |
Hb_011609_170 |
0.0688670271 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 4 |
Hb_000329_130 |
0.0723270799 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 5 |
Hb_002675_040 |
0.0735986995 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 6 |
Hb_005408_020 |
0.0790845009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001900_120 |
0.079390485 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 8 |
Hb_147737_010 |
0.0822820612 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000331_190 |
0.0823797238 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 10 |
Hb_033594_020 |
0.0826021397 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 11 |
Hb_000120_620 |
0.0830272395 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 12 |
Hb_125092_020 |
0.0833800967 |
- |
- |
PREDICTED: nucleolin 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000120_510 |
0.0838831311 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002307_350 |
0.0838945345 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 15 |
Hb_000343_210 |
0.0865027075 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000531_090 |
0.0869036891 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 17 |
Hb_004920_060 |
0.0876371643 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 18 |
Hb_030827_030 |
0.0885756934 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 19 |
Hb_009151_020 |
0.0894924385 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 20 |
Hb_001489_070 |
0.0897918595 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |