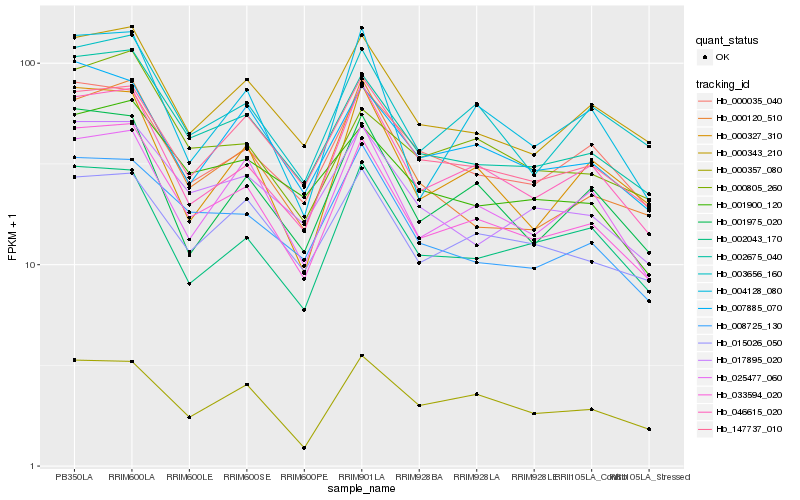
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033594_020 |
0.0 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 2 |
Hb_002675_040 |
0.0447412082 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 3 |
Hb_046615_020 |
0.0667299987 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 4 |
Hb_000343_210 |
0.0740628668 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 5 |
Hb_007885_070 |
0.0776826412 |
- |
- |
cleavage and polyadenylation specificity factor, putative [Ricinus communis] |
| 6 |
Hb_003656_160 |
0.0792056486 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein B456_013G226400 [Gossypium raimondii] |
| 7 |
Hb_017895_020 |
0.0793447605 |
- |
- |
Vacuolar protein sorting protein, putative [Ricinus communis] |
| 8 |
Hb_025477_060 |
0.080332529 |
- |
- |
PREDICTED: calcium-dependent protein kinase 26-like [Jatropha curcas] |
| 9 |
Hb_001975_020 |
0.0807468908 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 10 |
Hb_008725_130 |
0.0826021397 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 11 |
Hb_147737_010 |
0.0847544501 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000805_260 |
0.0852215994 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002043_170 |
0.0866451414 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 14 |
Hb_000035_040 |
0.0866453766 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 15 |
Hb_000357_080 |
0.0867028011 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59200, chloroplastic [Jatropha curcas] |
| 16 |
Hb_015026_050 |
0.0871783963 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_000120_510 |
0.0874590353 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001900_120 |
0.0879649347 |
- |
- |
PREDICTED: uncharacterized protein LOC105632666 [Jatropha curcas] |
| 19 |
Hb_004128_080 |
0.0882843419 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator-like APRR7 [Jatropha curcas] |
| 20 |
Hb_000327_310 |
0.0893235889 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |