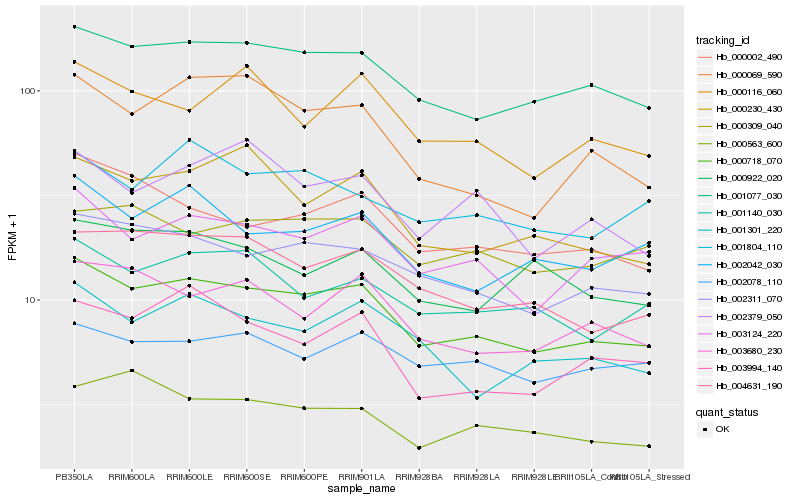
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000718_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001077_030 |
0.0510046117 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002311_070 |
0.057980053 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 4 |
Hb_003124_220 |
0.0660164667 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 93 [Jatropha curcas] |
| 5 |
Hb_004631_190 |
0.0699906408 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 6 |
Hb_003680_230 |
0.0744449023 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 7 |
Hb_001140_030 |
0.0756336374 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 8 |
Hb_000002_490 |
0.0762035772 |
- |
- |
PREDICTED: uncharacterized protein LOC105632095 [Jatropha curcas] |
| 9 |
Hb_002042_030 |
0.0767492253 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 10 |
Hb_001301_220 |
0.0778541628 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 11 |
Hb_000230_430 |
0.0798121289 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000116_060 |
0.0825768435 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 13 |
Hb_001804_110 |
0.084252376 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 14 |
Hb_003994_140 |
0.0844726833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002078_110 |
0.0848800086 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 16 |
Hb_000069_590 |
0.0856316594 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 17 |
Hb_000922_020 |
0.0865586365 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000309_040 |
0.0876912591 |
- |
- |
PREDICTED: protein farnesyltransferase subunit beta isoform X1 [Jatropha curcas] |
| 19 |
Hb_000563_600 |
0.0881519606 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 20 |
Hb_002379_050 |
0.088164842 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |