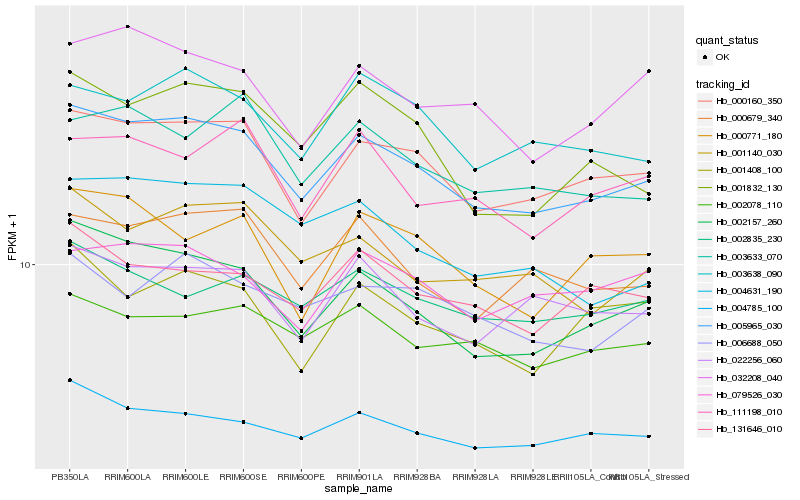
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005965_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 2 |
Hb_000160_350 |
0.0422792461 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 3 |
Hb_002157_260 |
0.0534358683 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 4 |
Hb_004785_100 |
0.0621118909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000679_340 |
0.0644238238 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003638_090 |
0.0674963373 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 7 |
Hb_022256_060 |
0.06833026 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 8 |
Hb_001140_030 |
0.0703095098 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 9 |
Hb_131646_010 |
0.0714721167 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_032208_040 |
0.071512517 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 11 |
Hb_002078_110 |
0.0718450146 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 12 |
Hb_006688_050 |
0.0730689096 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX15 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003633_070 |
0.0734425135 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 14 |
Hb_001832_130 |
0.0746961501 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 15 |
Hb_079526_030 |
0.0750455426 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 16 |
Hb_002835_230 |
0.0757294417 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000771_180 |
0.0766991096 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 18 |
Hb_004631_190 |
0.076959815 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 19 |
Hb_111198_010 |
0.0777435332 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001408_100 |
0.0781143134 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |