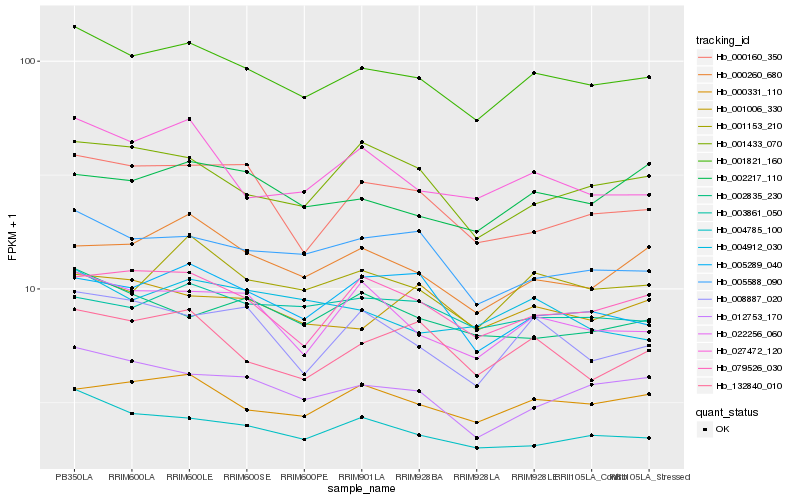
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001821_160 |
0.0 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 2 |
Hb_004785_100 |
0.0556876751 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000331_110 |
0.0633956663 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 4 |
Hb_079526_030 |
0.0634148181 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 5 |
Hb_012753_170 |
0.0637866842 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_005588_090 |
0.0656537968 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 7 |
Hb_022256_060 |
0.0671428136 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 8 |
Hb_001006_330 |
0.0710939842 |
- |
- |
sodium/hydrogen exchanger plant, putative [Ricinus communis] |
| 9 |
Hb_000260_680 |
0.0714024721 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 10 |
Hb_027472_120 |
0.0728902029 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001433_070 |
0.0729656641 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 12 |
Hb_002835_230 |
0.0729916328 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002217_110 |
0.0733502748 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 14 |
Hb_003861_050 |
0.0734321329 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 15 |
Hb_132840_010 |
0.0737765519 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000160_350 |
0.076029585 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 17 |
Hb_001153_210 |
0.0760934603 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008887_020 |
0.0764559904 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 19 |
Hb_005289_040 |
0.0767102978 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 20 |
Hb_004912_030 |
0.0770291696 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |