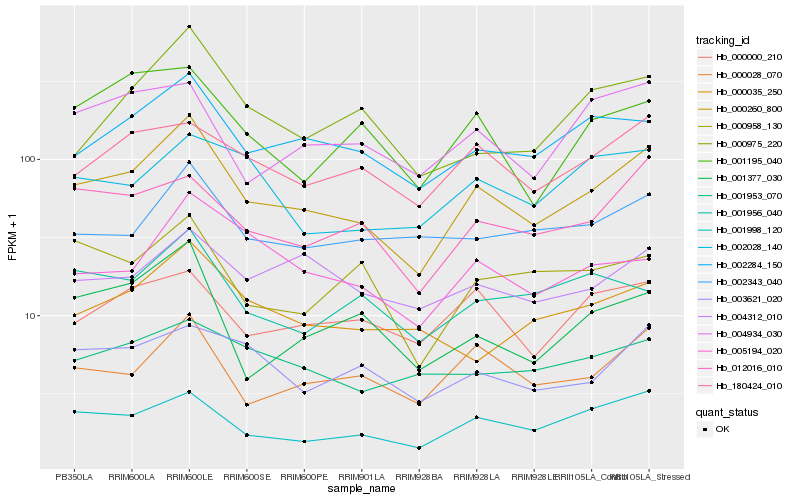
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_800 |
0.0 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 [Populus euphratica] |
| 2 |
Hb_001998_120 |
0.1094469332 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 3 |
Hb_002284_150 |
0.114153948 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 4 |
Hb_000028_070 |
0.1147811931 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 5 |
Hb_012016_010 |
0.1255683101 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 6, mitochondrial [Jatropha curcas] |
| 6 |
Hb_180424_010 |
0.1301980877 |
- |
- |
Charged multivesicular body protein, putative [Ricinus communis] |
| 7 |
Hb_001377_030 |
0.1323319582 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 8 |
Hb_002343_040 |
0.1345259297 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 2, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_005194_020 |
0.1353307921 |
- |
- |
PREDICTED: UPF0553 protein-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_003621_020 |
0.1360119734 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 11 |
Hb_000000_210 |
0.1360785496 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004934_030 |
0.1367808907 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 13 |
Hb_000975_220 |
0.1369874255 |
- |
- |
Histone H2B [Medicago truncatula] |
| 14 |
Hb_004312_010 |
0.1390234208 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000035_250 |
0.1398511854 |
- |
- |
PREDICTED: uncharacterized protein LOC105637544 [Jatropha curcas] |
| 16 |
Hb_001956_040 |
0.1426123627 |
- |
- |
PREDICTED: PGR5-like protein 1B, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_001195_040 |
0.1428223343 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 18 |
Hb_001953_070 |
0.1434436844 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002028_140 |
0.1443906326 |
transcription factor |
TF Family: Tify |
JAZ11 [Hevea brasiliensis] |
| 20 |
Hb_000958_130 |
0.1449520269 |
- |
- |
PREDICTED: uncharacterized protein LOC105628716 isoform X1 [Jatropha curcas] |