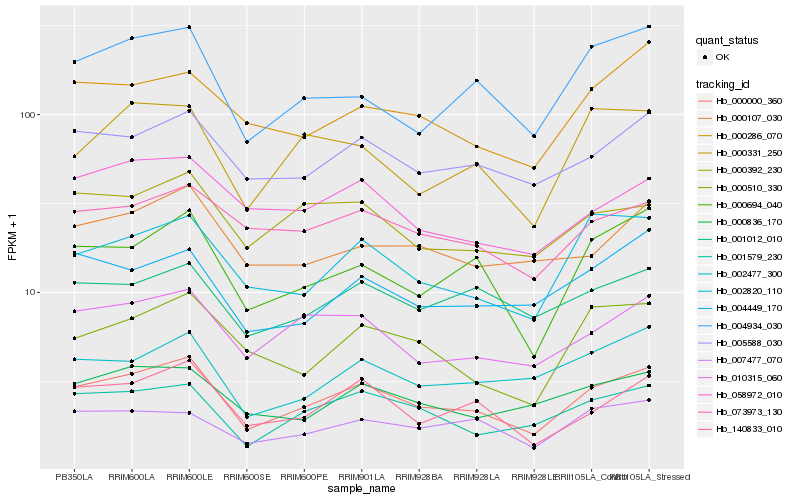
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_360 |
0.0 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 2 |
Hb_140833_010 |
0.0864225505 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 3 |
Hb_000694_040 |
0.0871975165 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 4 |
Hb_001579_230 |
0.0889366499 |
- |
- |
PREDICTED: uncharacterized protein LOC105644968 [Jatropha curcas] |
| 5 |
Hb_005588_030 |
0.0892460478 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 6 |
Hb_010315_060 |
0.0894576581 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 7 |
Hb_002820_110 |
0.0915782419 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 8 |
Hb_004934_030 |
0.0958377368 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 [Cucumis melo] |
| 9 |
Hb_000836_170 |
0.1003709474 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 10 |
Hb_007477_070 |
0.1010064427 |
- |
- |
hypothetical protein JCGZ_17593 [Jatropha curcas] |
| 11 |
Hb_000510_330 |
0.1014360457 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 12 |
Hb_073973_130 |
0.1016200235 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_002477_300 |
0.1016808998 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000331_250 |
0.1030999774 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 1 [Vitis vinifera] |
| 15 |
Hb_000107_030 |
0.103403219 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_058972_010 |
0.1036002264 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004449_170 |
0.1037160313 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000392_230 |
0.1041378933 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000286_070 |
0.104632307 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 20 |
Hb_001012_010 |
0.105085087 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |