| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
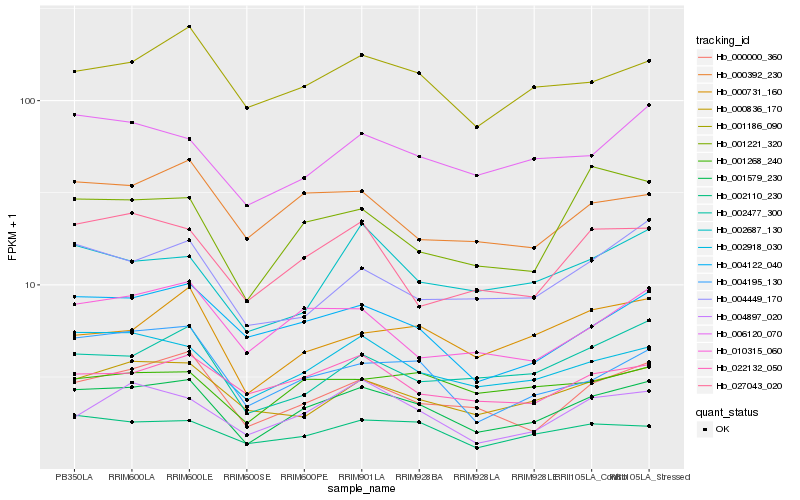
Hb_001579_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644968 [Jatropha curcas] |
| 2 |
Hb_002918_030 |
0.0836152139 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000000_360 |
0.0889366499 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 4 |
Hb_004122_040 |
0.0973213608 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 5 |
Hb_002110_230 |
0.0985628794 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 6 |
Hb_000836_170 |
0.0987347287 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 7 |
Hb_004195_130 |
0.0994209555 |
- |
- |
PREDICTED: thioredoxin domain-containing protein PLP3B [Sesamum indicum] |
| 8 |
Hb_006120_070 |
0.1014332231 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 9 |
Hb_001186_090 |
0.1035891408 |
- |
- |
PREDICTED: nascent polypeptide-associated complex subunit alpha-like protein 2 [Jatropha curcas] |
| 10 |
Hb_004897_020 |
0.1036340551 |
- |
- |
PREDICTED: (-)-germacrene D synthase-like [Populus euphratica] |
| 11 |
Hb_027043_020 |
0.104199382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_010315_060 |
0.1059144546 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 13 |
Hb_002477_300 |
0.1062406116 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001221_320 |
0.1063899629 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_022132_050 |
0.1073669979 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002687_130 |
0.1077985184 |
- |
- |
PREDICTED: uncharacterized protein LOC105631153 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004449_170 |
0.1081945746 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000731_160 |
0.1082398591 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 19 |
Hb_000392_230 |
0.108438044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001268_240 |
0.1092802436 |
- |
- |
PREDICTED: uncharacterized protein LOC105630174 isoform X2 [Jatropha curcas] |