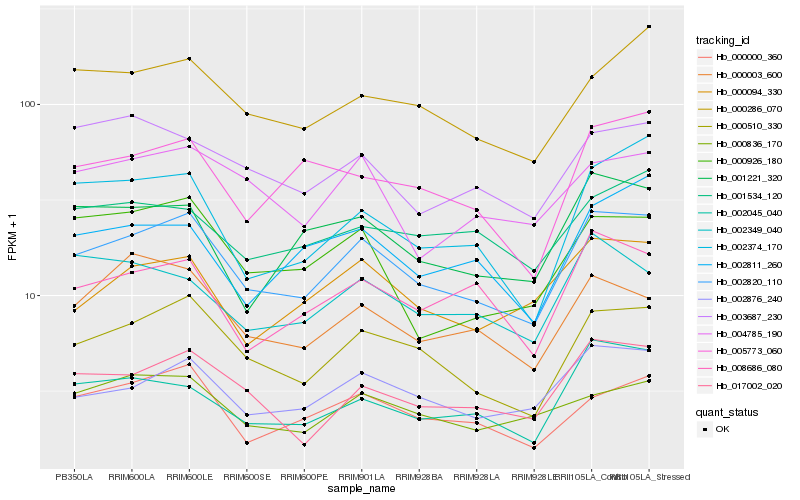
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002820_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 2 |
Hb_000510_330 |
0.0572723247 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 3 |
Hb_002876_240 |
0.0731731695 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_017002_020 |
0.0885254044 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 5 |
Hb_000000_360 |
0.0915782419 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 6 |
Hb_008686_080 |
0.0966491932 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5A-like [Jatropha curcas] |
| 7 |
Hb_000094_330 |
0.09702844 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 8 |
Hb_000286_070 |
0.0974507837 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 9 |
Hb_001221_320 |
0.0998089839 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000836_170 |
0.1017225164 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 11 |
Hb_000003_600 |
0.1029574274 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 12 |
Hb_002045_040 |
0.1039940587 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005773_060 |
0.1045770331 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 14 |
Hb_004785_190 |
0.1048029831 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 15 |
Hb_000926_180 |
0.1071163877 |
- |
- |
proline synthetase associated protein, putative [Ricinus communis] |
| 16 |
Hb_001534_120 |
0.1073977941 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 17 |
Hb_002374_170 |
0.1076246065 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |
| 18 |
Hb_003687_230 |
0.1077778578 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 19 |
Hb_002811_260 |
0.1078437294 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 20 |
Hb_002349_040 |
0.1081075868 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |