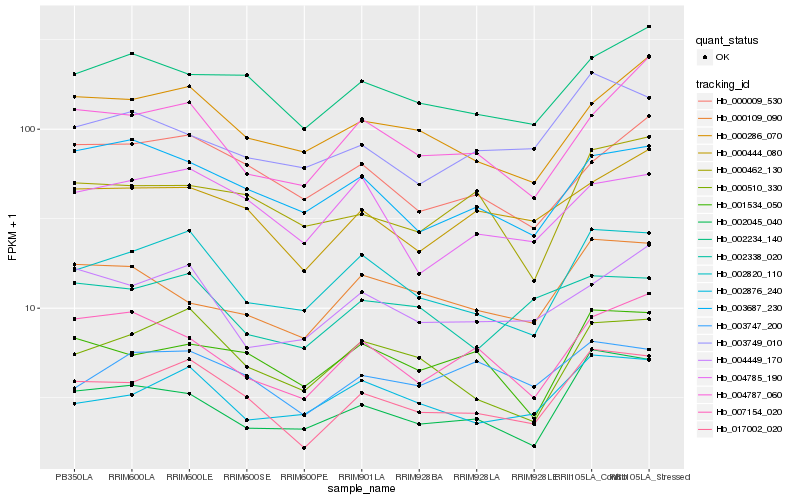
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017002_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 2 |
Hb_002820_110 |
0.0885254044 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 3 |
Hb_000510_330 |
0.1058431426 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 4 |
Hb_000444_080 |
0.1084917633 |
- |
- |
hypothetical protein POPTR_0019s10500g [Populus trichocarpa] |
| 5 |
Hb_002045_040 |
0.1090562074 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001534_050 |
0.1121296754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642299 [Jatropha curcas] |
| 7 |
Hb_002876_240 |
0.1123734737 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_002234_140 |
0.1149371071 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 9 |
Hb_000286_070 |
0.1203913863 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 10 |
Hb_003747_200 |
0.1233908418 |
- |
- |
PREDICTED: DNA cross-link repair protein SNM1 [Jatropha curcas] |
| 11 |
Hb_000462_130 |
0.1237931299 |
- |
- |
hypothetical protein CICLE_v10009355mg [Citrus clementina] |
| 12 |
Hb_004785_190 |
0.1252172668 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 13 |
Hb_007154_020 |
0.1256439383 |
- |
- |
- |
| 14 |
Hb_003749_010 |
0.1267056088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000009_530 |
0.1269650973 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 16 |
Hb_003687_230 |
0.1269674338 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 17 |
Hb_000109_090 |
0.1273298113 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 18 |
Hb_004449_170 |
0.1274174105 |
- |
- |
PREDICTED: uncharacterized protein LOC105631285 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004787_060 |
0.1290353163 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 20 |
Hb_002338_020 |
0.1290585027 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |