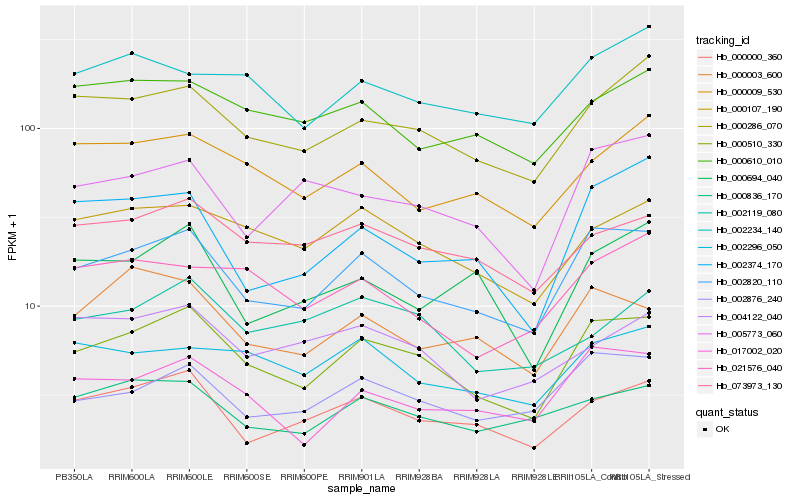
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000510_330 |
0.0 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 2 |
Hb_002820_110 |
0.0572723247 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 3 |
Hb_000286_070 |
0.0996840314 |
- |
- |
PREDICTED: 60S ribosomal protein L13-1-like [Jatropha curcas] |
| 4 |
Hb_000000_360 |
0.1014360457 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 5 |
Hb_000107_190 |
0.1034198829 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 6 |
Hb_017002_020 |
0.1058431426 |
- |
- |
PREDICTED: uncharacterized protein LOC105650411 [Jatropha curcas] |
| 7 |
Hb_002876_240 |
0.1061238604 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_073973_130 |
0.1127699236 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 9 |
Hb_000836_170 |
0.1170170561 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 10 |
Hb_000003_600 |
0.1184947255 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 11 |
Hb_000009_530 |
0.1196172388 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 12 |
Hb_005773_060 |
0.1203220349 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 13 |
Hb_002296_050 |
0.1212817867 |
- |
- |
hypothetical protein AMTR_s00001p00247600 [Amborella trichopoda] |
| 14 |
Hb_004122_040 |
0.1238509921 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_000610_010 |
0.1248779902 |
- |
- |
PREDICTED: SKP1-like protein 1B [Musa acuminata subsp. malaccensis] |
| 16 |
Hb_021576_040 |
0.1250437883 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002119_080 |
0.1261644665 |
- |
- |
PREDICTED: 54S ribosomal protein L24, mitochondrial [Jatropha curcas] |
| 18 |
Hb_002234_140 |
0.1267213125 |
- |
- |
DNA-directed RNA polymerase II family protein [Populus trichocarpa] |
| 19 |
Hb_000694_040 |
0.127674189 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_002374_170 |
0.1277831256 |
- |
- |
20S proteasome beta subunit C1 [Hevea brasiliensis] |