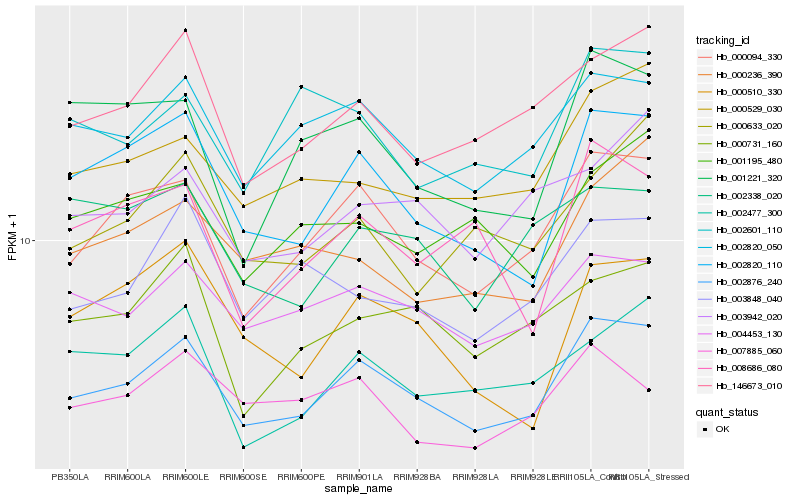
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_240 |
0.0 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000094_330 |
0.0601554807 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_002820_110 |
0.0731731695 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 4 |
Hb_002820_050 |
0.0794443507 |
- |
- |
PREDICTED: putative glutathione-specific gamma-glutamylcyclotransferase 2 [Jatropha curcas] |
| 5 |
Hb_004453_130 |
0.0850677112 |
- |
- |
PREDICTED: tRNA (cytosine-5-)-methyltransferase isoform X1 [Jatropha curcas] |
| 6 |
Hb_146673_010 |
0.085456078 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 7 |
Hb_000529_030 |
0.0910178525 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_003848_040 |
0.1011032045 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_000731_160 |
0.1016322919 |
- |
- |
PREDICTED: adenosine deaminase-like protein [Jatropha curcas] |
| 10 |
Hb_003942_020 |
0.1020007013 |
- |
- |
PREDICTED: multiple RNA-binding domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002477_300 |
0.1024964366 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_001221_320 |
0.1055385136 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000510_330 |
0.1061238604 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 14 |
Hb_008686_080 |
0.1064905564 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5A-like [Jatropha curcas] |
| 15 |
Hb_001195_480 |
0.1067329306 |
- |
- |
cop9 complex subunit, putative [Ricinus communis] |
| 16 |
Hb_000236_390 |
0.1075656878 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH1 [Jatropha curcas] |
| 17 |
Hb_000633_020 |
0.1086378995 |
- |
- |
PREDICTED: uncharacterized protein LOC105630002 [Jatropha curcas] |
| 18 |
Hb_007885_060 |
0.108939203 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 19 |
Hb_002601_110 |
0.1092421508 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 20 |
Hb_002338_020 |
0.1098502091 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR34A [Jatropha curcas] |