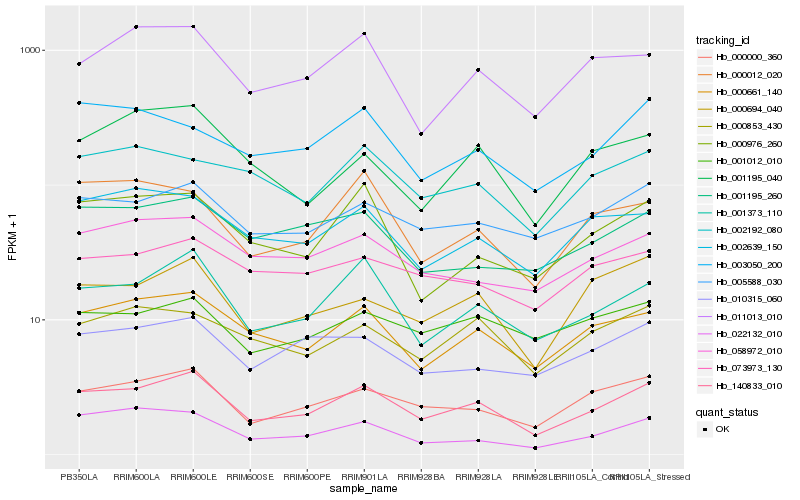
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140833_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 2 |
Hb_001373_110 |
0.0810715545 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 3 |
Hb_000000_360 |
0.0864225505 |
- |
- |
PREDICTED: methyltransferase-like protein 2 [Populus euphratica] |
| 4 |
Hb_000661_140 |
0.0892945629 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005588_030 |
0.1039558795 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 6 |
Hb_000694_040 |
0.1084767212 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_011013_010 |
0.111351288 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 8 |
Hb_002639_150 |
0.1126337997 |
- |
- |
- |
| 9 |
Hb_000853_430 |
0.1136095168 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 10 |
Hb_058972_010 |
0.1141221655 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 11 |
Hb_010315_060 |
0.1157767852 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 12 |
Hb_000976_260 |
0.1163215339 |
- |
- |
- |
| 13 |
Hb_000012_020 |
0.1182839382 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 14 |
Hb_002192_080 |
0.1195842232 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 15 |
Hb_022132_010 |
0.1196665592 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_001195_260 |
0.1209632429 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 17 |
Hb_073973_130 |
0.1226529142 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_003050_200 |
0.1227879643 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 19 |
Hb_001195_040 |
0.1229670566 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 20 |
Hb_001012_010 |
0.1233056879 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |