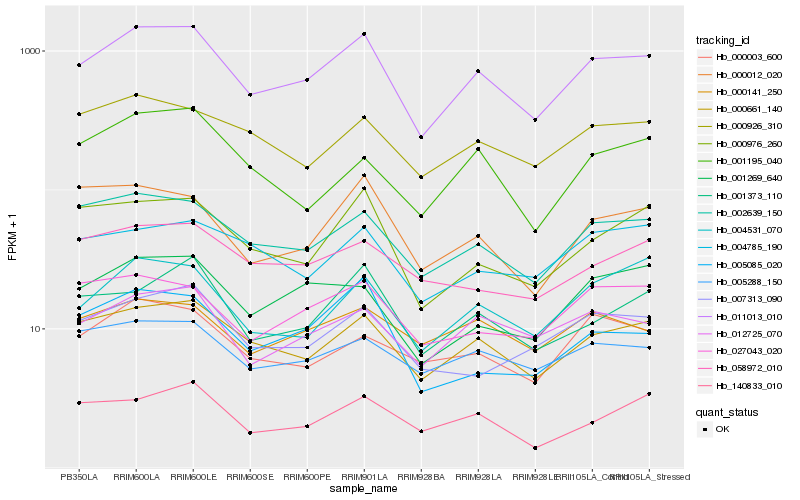
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011013_010 |
0.0 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 2 |
Hb_000661_140 |
0.0703024416 |
- |
- |
PREDICTED: uncharacterized protein LOC105648317 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002639_150 |
0.0817730005 |
- |
- |
- |
| 4 |
Hb_012725_070 |
0.0922102354 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005288_150 |
0.1019519763 |
- |
- |
PREDICTED: casein kinase I-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_001373_110 |
0.1020139788 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 7 |
Hb_004531_070 |
0.1057297541 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX19-like [Jatropha curcas] |
| 8 |
Hb_000976_260 |
0.1059465837 |
- |
- |
- |
| 9 |
Hb_000926_310 |
0.1060732301 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 10 |
Hb_001269_640 |
0.1075553351 |
- |
- |
hypothetical protein CISIN_1g032720mg [Citrus sinensis] |
| 11 |
Hb_027043_020 |
0.1075888216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007313_090 |
0.110219507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000012_020 |
0.110746422 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 14 |
Hb_140833_010 |
0.111351288 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 15 |
Hb_000141_250 |
0.1115748116 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X3 [Jatropha curcas] |
| 16 |
Hb_000003_600 |
0.1126663032 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1 [Jatropha curcas] |
| 17 |
Hb_004785_190 |
0.1150738201 |
- |
- |
PHD finger family protein [Populus trichocarpa] |
| 18 |
Hb_005085_020 |
0.1160187024 |
- |
- |
hypothetical protein B456_001G193200 [Gossypium raimondii] |
| 19 |
Hb_058972_010 |
0.1162832074 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001195_040 |
0.120147058 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |