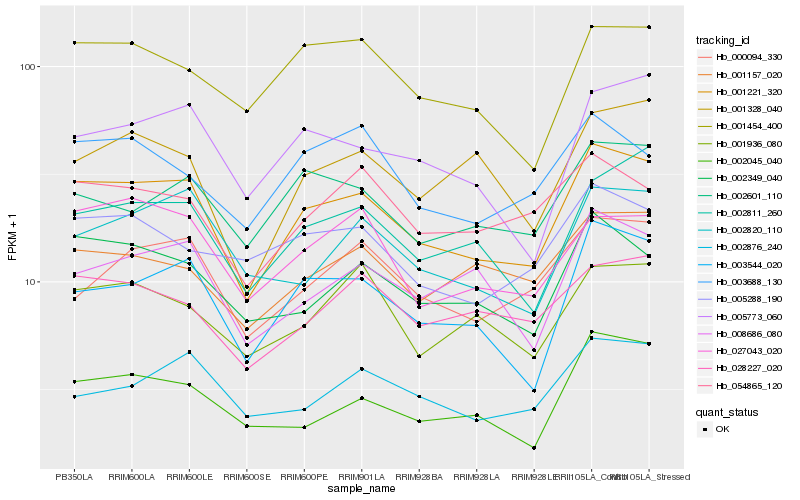
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_320 |
0.0 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003544_020 |
0.0790438806 |
- |
- |
PREDICTED: elongin-A [Jatropha curcas] |
| 3 |
Hb_002349_040 |
0.088615472 |
- |
- |
PREDICTED: RNA pseudouridine synthase 5 [Jatropha curcas] |
| 4 |
Hb_002811_260 |
0.0926089621 |
- |
- |
PREDICTED: PITH domain-containing protein At3g04780 [Jatropha curcas] |
| 5 |
Hb_005773_060 |
0.0955915612 |
- |
- |
defender against cell death, putative [Ricinus communis] |
| 6 |
Hb_008686_080 |
0.0959285693 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5A-like [Jatropha curcas] |
| 7 |
Hb_001454_400 |
0.0971239121 |
- |
- |
PREDICTED: dynein light chain 1, cytoplasmic [Jatropha curcas] |
| 8 |
Hb_002601_110 |
0.0982864129 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 9 |
Hb_027043_020 |
0.0984801142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002820_110 |
0.0998089839 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 11 |
Hb_000094_330 |
0.1001125752 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 4 [Jatropha curcas] |
| 12 |
Hb_028227_020 |
0.1018983778 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB isoform X1 [Jatropha curcas] |
| 13 |
Hb_054865_120 |
0.1024902473 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 14 |
Hb_001157_020 |
0.1037183569 |
- |
- |
PREDICTED: uncharacterized protein LOC105649844 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002045_040 |
0.1042365939 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001936_080 |
0.104581344 |
- |
- |
PREDICTED: RNA-binding protein 48-like [Jatropha curcas] |
| 17 |
Hb_001328_040 |
0.1048345578 |
- |
- |
PREDICTED: uncharacterized protein LOC105636146 [Jatropha curcas] |
| 18 |
Hb_003688_130 |
0.105067947 |
- |
- |
phosphoserine phosphatase, putative [Ricinus communis] |
| 19 |
Hb_005288_190 |
0.1051997716 |
- |
- |
PREDICTED: exportin-4 [Jatropha curcas] |
| 20 |
Hb_002876_240 |
0.1055385136 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |