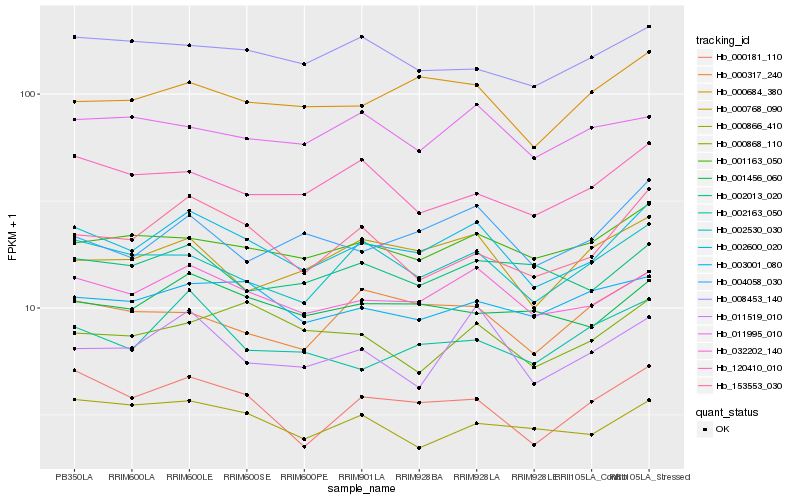
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002600_020 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_032202_140 |
0.0443292594 |
- |
- |
PREDICTED: flap endonuclease GEN-like 1 [Jatropha curcas] |
| 3 |
Hb_000181_110 |
0.072756902 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 4 |
Hb_002530_030 |
0.0741453167 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 5 |
Hb_001163_050 |
0.0747297394 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 6 |
Hb_000684_380 |
0.0755294014 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 7 |
Hb_153553_030 |
0.0762965846 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 8 |
Hb_001456_060 |
0.0772820245 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 9 |
Hb_011519_010 |
0.0789704771 |
- |
- |
PREDICTED: uncharacterized protein LOC105645310 [Jatropha curcas] |
| 10 |
Hb_003001_080 |
0.0794869367 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 11 |
Hb_004058_030 |
0.0831471605 |
- |
- |
PREDICTED: stomatin-like protein 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000768_090 |
0.0831645579 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase isoform X1 [Jatropha curcas] |
| 13 |
Hb_002013_020 |
0.0847237228 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 14 |
Hb_120410_010 |
0.0855696472 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 15 |
Hb_008453_140 |
0.085818453 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 16 |
Hb_002163_050 |
0.0873848329 |
- |
- |
hypothetical protein POPTR_0014s15490g [Populus trichocarpa] |
| 17 |
Hb_000866_410 |
0.0875398616 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 18 |
Hb_011995_010 |
0.0883287991 |
- |
- |
dc50, putative [Ricinus communis] |
| 19 |
Hb_000868_110 |
0.0892377909 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 20 |
Hb_000317_240 |
0.0897081617 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |