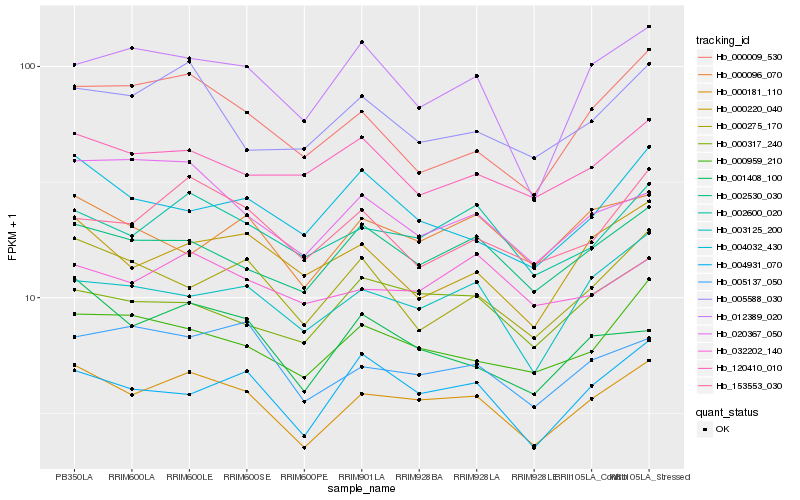
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637521 [Jatropha curcas] |
| 2 |
Hb_002600_020 |
0.072756902 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_002530_030 |
0.0756129032 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 4 |
Hb_000959_210 |
0.0847536791 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 27 [Jatropha curcas] |
| 5 |
Hb_003125_200 |
0.084974104 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 30 [Jatropha curcas] |
| 6 |
Hb_004931_070 |
0.0881469932 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000317_240 |
0.0887341273 |
- |
- |
PREDICTED: protein DYAD [Jatropha curcas] |
| 8 |
Hb_005137_050 |
0.0899102168 |
- |
- |
PREDICTED: nuclear pore complex protein NUP58 [Jatropha curcas] |
| 9 |
Hb_001408_100 |
0.0911829775 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 10 |
Hb_000220_040 |
0.0922220917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005588_030 |
0.0926416354 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 14 homolog [Eucalyptus grandis] |
| 12 |
Hb_012389_020 |
0.0944953143 |
- |
- |
hypothetical protein POPTR_0006s05190g [Populus trichocarpa] |
| 13 |
Hb_153553_030 |
0.0946591874 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 14 |
Hb_000096_070 |
0.0947406301 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 15 |
Hb_020367_050 |
0.0954752126 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_000275_170 |
0.0955820714 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 17 |
Hb_032202_140 |
0.0956853894 |
- |
- |
PREDICTED: flap endonuclease GEN-like 1 [Jatropha curcas] |
| 18 |
Hb_000009_530 |
0.0967995974 |
- |
- |
PREDICTED: uncharacterized protein LOC105639628 [Jatropha curcas] |
| 19 |
Hb_120410_010 |
0.0978092835 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 20 |
Hb_004032_430 |
0.0980721415 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |