| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
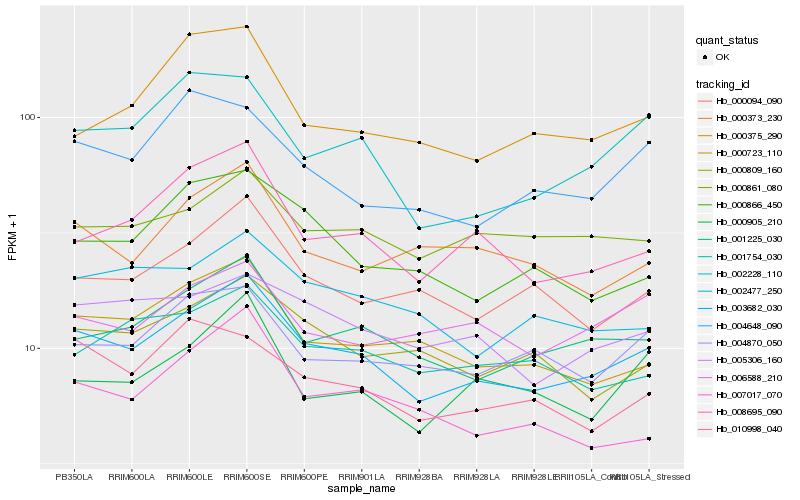
Hb_003682_030 |
0.0 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 2 |
Hb_001225_030 |
0.0695719182 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 3 |
Hb_008695_090 |
0.079739367 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 4 |
Hb_000723_110 |
0.0822610974 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 5 |
Hb_005306_160 |
0.0824516902 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1-like [Jatropha curcas] |
| 6 |
Hb_004870_050 |
0.0834554296 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 7 |
Hb_000094_090 |
0.0836646667 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002228_110 |
0.0858658528 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 9 |
Hb_000905_210 |
0.0875330162 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |
| 10 |
Hb_000809_160 |
0.0882316386 |
- |
- |
hypothetical protein JCGZ_12355 [Jatropha curcas] |
| 11 |
Hb_000861_080 |
0.0889984902 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 12 |
Hb_002477_250 |
0.0895888561 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 13 |
Hb_001754_030 |
0.0901662686 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 14 |
Hb_004648_090 |
0.0909517874 |
- |
- |
peroxisomal biogenesis factor, putative [Ricinus communis] |
| 15 |
Hb_000866_450 |
0.0912347302 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_010998_040 |
0.091398932 |
transcription factor |
TF Family: MYB |
myb3r3, putative [Ricinus communis] |
| 17 |
Hb_000373_230 |
0.0918567558 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 18 |
Hb_006588_210 |
0.0926335562 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 19 |
Hb_007017_070 |
0.0951612539 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000375_290 |
0.0956224758 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |