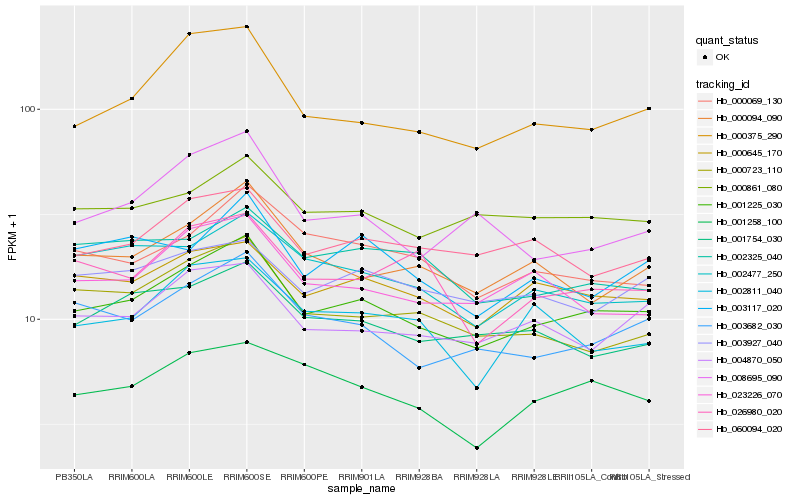
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001225_030 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 2 |
Hb_003682_030 |
0.0695719182 |
- |
- |
hypothetical protein CICLE_v10020876mg [Citrus clementina] |
| 3 |
Hb_000375_290 |
0.0695810768 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000645_170 |
0.0713357771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004870_050 |
0.0754033258 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 6 |
Hb_000094_090 |
0.0760781069 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001754_030 |
0.0792437898 |
- |
- |
PREDICTED: ubiquitin-like-specific protease ESD4 [Jatropha curcas] |
| 8 |
Hb_000861_080 |
0.0793893763 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 9 |
Hb_060094_020 |
0.0794463851 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 10 |
Hb_023226_070 |
0.0796229517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000069_130 |
0.0807388706 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000723_110 |
0.0811979725 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 13 |
Hb_001258_100 |
0.081250939 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_002477_250 |
0.0833360891 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 15 |
Hb_003927_040 |
0.0834564369 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 16 |
Hb_003117_020 |
0.083477091 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE6 [Jatropha curcas] |
| 17 |
Hb_026980_020 |
0.0875941406 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 18 |
Hb_008695_090 |
0.0883747197 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 19 |
Hb_002325_040 |
0.0892752163 |
- |
- |
PREDICTED: uncharacterized protein LOC105645422 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002811_040 |
0.0902621986 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |