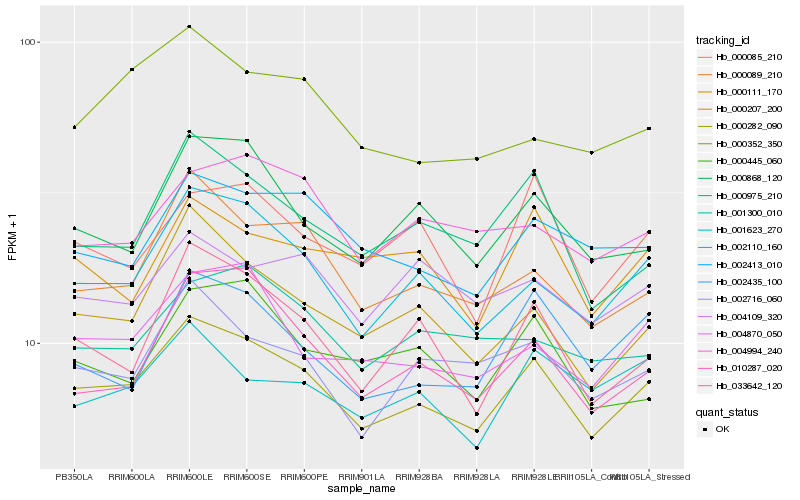
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000282_090 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 2 |
Hb_002110_160 |
0.0582814944 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000207_200 |
0.070479032 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_004870_050 |
0.0720055823 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP3 [Jatropha curcas] |
| 5 |
Hb_000089_210 |
0.0721802638 |
- |
- |
unknown [Medicago truncatula] |
| 6 |
Hb_002716_060 |
0.0739423292 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 7 |
Hb_002413_010 |
0.0744801923 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000868_120 |
0.0750347854 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 9 |
Hb_033642_120 |
0.0757684429 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 10 |
Hb_000975_210 |
0.0768837927 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 11 |
Hb_002435_100 |
0.0785523621 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 12 |
Hb_010287_020 |
0.0792922215 |
- |
- |
CCR4-NOT transcription complex subunit, putative [Ricinus communis] |
| 13 |
Hb_000111_170 |
0.0815752837 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000085_210 |
0.0822186826 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 15 |
Hb_004994_240 |
0.0825550308 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 16 |
Hb_001623_270 |
0.0825964744 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 17 |
Hb_004109_320 |
0.0834406405 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 18 |
Hb_000445_060 |
0.0836353321 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001300_010 |
0.084336131 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000352_350 |
0.0853904646 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |