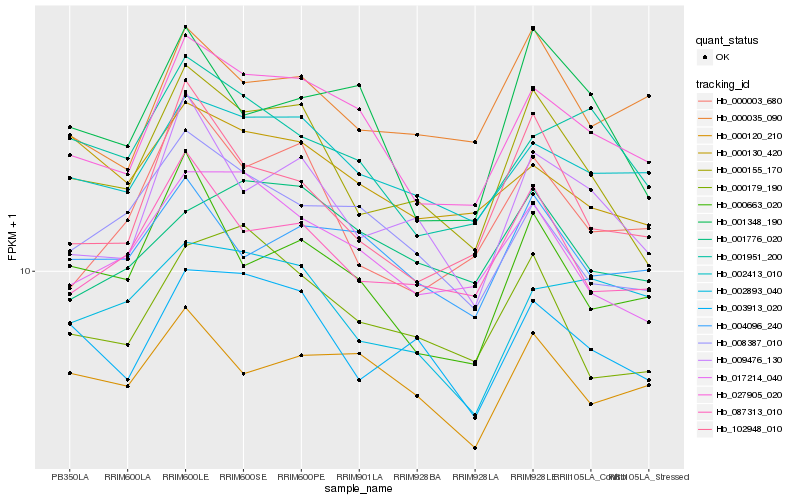
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027905_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 2 |
Hb_002413_010 |
0.0661414913 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_017214_040 |
0.0846568006 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase COP1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001951_200 |
0.0875257842 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_004096_240 |
0.0917505691 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 6 |
Hb_008387_010 |
0.0929631972 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 7 |
Hb_000120_210 |
0.09308094 |
- |
- |
PREDICTED: uncharacterized protein LOC105630189 [Jatropha curcas] |
| 8 |
Hb_009476_130 |
0.0975400909 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 9 |
Hb_087313_010 |
0.0978751941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000663_020 |
0.0987464811 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_000155_170 |
0.0989569026 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 12 |
Hb_000035_090 |
0.0997252433 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 13 |
Hb_001776_020 |
0.0998181097 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 14 |
Hb_000130_420 |
0.1021562256 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 15 |
Hb_000003_680 |
0.1025064431 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 16 |
Hb_102948_010 |
0.1027542981 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001348_190 |
0.1031799984 |
- |
- |
PREDICTED: chlorophyllide a oxygenase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_003913_020 |
0.1032595633 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At5g50170 [Jatropha curcas] |
| 19 |
Hb_000179_190 |
0.1036893674 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 isoform X1 [Vitis vinifera] |
| 20 |
Hb_002893_040 |
0.1036981881 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |