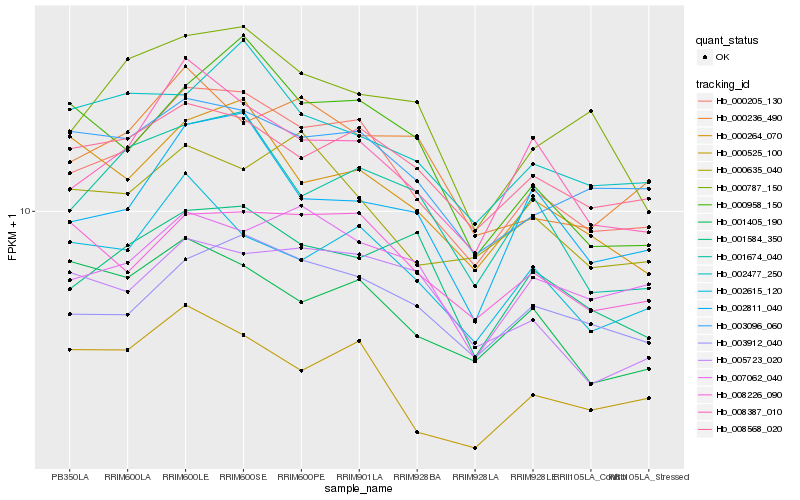
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000205_130 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_008387_010 |
0.0681027137 |
- |
- |
SMAD/FHA domain-containing protein isoform 3 [Theobroma cacao] |
| 3 |
Hb_003912_040 |
0.0717552568 |
- |
- |
hypothetical protein CISIN_1g005153mg [Citrus sinensis] |
| 4 |
Hb_008568_020 |
0.071784273 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 5 |
Hb_003096_060 |
0.072614092 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 6 |
Hb_002615_120 |
0.0737795442 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000525_100 |
0.075873568 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 8 |
Hb_008226_090 |
0.0768643061 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001674_040 |
0.0772952196 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 23 [Jatropha curcas] |
| 10 |
Hb_002811_040 |
0.0783992214 |
- |
- |
5'->3' exoribonuclease, putative [Ricinus communis] |
| 11 |
Hb_007062_040 |
0.0804824729 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 12 |
Hb_000264_070 |
0.0815230592 |
- |
- |
PREDICTED: uncharacterized protein LOC105649513 [Jatropha curcas] |
| 13 |
Hb_002477_250 |
0.0835446754 |
- |
- |
DNA-directed RNA polymerase I 49 kDa polypeptide, putative [Ricinus communis] |
| 14 |
Hb_005723_020 |
0.0851020548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001405_190 |
0.0857915846 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_001584_350 |
0.0859176427 |
- |
- |
hypothetical protein JCGZ_23178 [Jatropha curcas] |
| 17 |
Hb_000787_150 |
0.08635025 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 18 |
Hb_000635_040 |
0.0867385051 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 19 |
Hb_000958_150 |
0.0868395578 |
- |
- |
Actin, putative [Ricinus communis] |
| 20 |
Hb_000236_490 |
0.0875361807 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |