| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
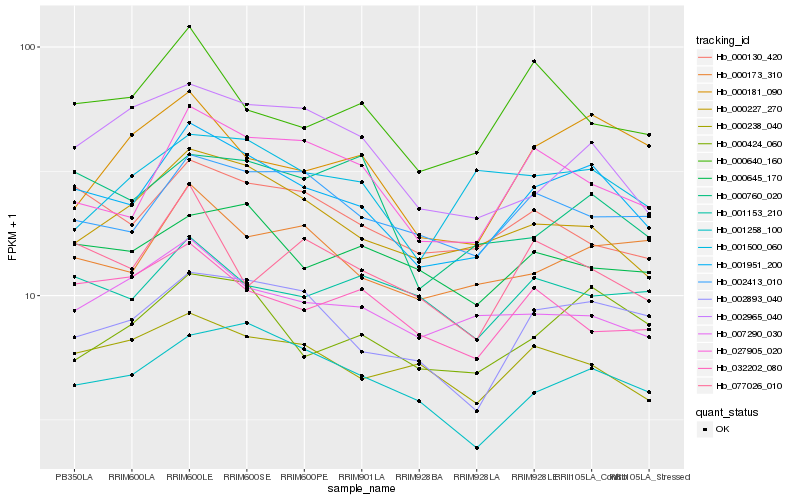
Hb_001951_200 |
0.0 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002893_040 |
0.0874883726 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 3 |
Hb_027905_020 |
0.0875257842 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 4 |
Hb_000227_270 |
0.0908686943 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002413_010 |
0.0921465717 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 6 |
Hb_000173_310 |
0.0926082956 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000424_060 |
0.0929991292 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 8 |
Hb_001258_100 |
0.0941566718 |
- |
- |
PREDICTED: probable starch synthase 4, chloroplastic/amyloplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_000130_420 |
0.0964012534 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_000238_040 |
0.1030281342 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 11 |
Hb_007290_030 |
0.1044705394 |
transcription factor |
TF Family: bZIP |
PREDICTED: uncharacterized protein LOC105629473 [Jatropha curcas] |
| 12 |
Hb_000645_170 |
0.1045564052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000640_160 |
0.1078801945 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 14 |
Hb_077026_010 |
0.109032535 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 2 [Jatropha curcas] |
| 15 |
Hb_001153_210 |
0.1115351945 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000760_020 |
0.1115856566 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_032202_080 |
0.1117545794 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001500_060 |
0.1120546873 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein ZFN-like [Jatropha curcas] |
| 19 |
Hb_002965_040 |
0.1125019242 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 20 |
Hb_000181_090 |
0.1129380269 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |