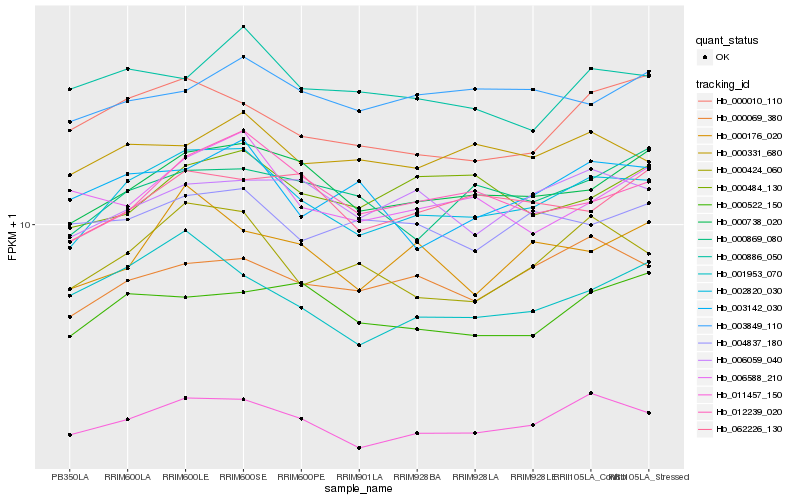
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002820_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 2 |
Hb_000738_020 |
0.0692309855 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_011457_150 |
0.0782065629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649946 [Jatropha curcas] |
| 4 |
Hb_012239_020 |
0.0789326804 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 5 |
Hb_000424_060 |
0.0791925553 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 6 |
Hb_000010_110 |
0.0880513462 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000069_380 |
0.0884302659 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000331_680 |
0.0910493066 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 9 |
Hb_062226_130 |
0.0918296806 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 4 [Jatropha curcas] |
| 10 |
Hb_001953_070 |
0.0925809778 |
- |
- |
PREDICTED: RNA pseudouridine synthase 7 isoform X2 [Jatropha curcas] |
| 11 |
Hb_006059_040 |
0.0933320764 |
transcription factor |
TF Family: SOH1 |
hypothetical protein PHAVU_007G063000g [Phaseolus vulgaris] |
| 12 |
Hb_000176_020 |
0.0938721196 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 13 |
Hb_000869_080 |
0.0943338256 |
- |
- |
PREDICTED: uncharacterized protein LOC105641084 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000886_050 |
0.0958189803 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 15 |
Hb_000522_150 |
0.095943081 |
- |
- |
PREDICTED: uncharacterized protein LOC105646670 [Jatropha curcas] |
| 16 |
Hb_006588_210 |
0.0965388281 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 17 |
Hb_004837_180 |
0.0974325895 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003142_030 |
0.098191335 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 14, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000484_130 |
0.0982431956 |
- |
- |
splicing factor, putative [Ricinus communis] |
| 20 |
Hb_003849_110 |
0.0983424098 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |