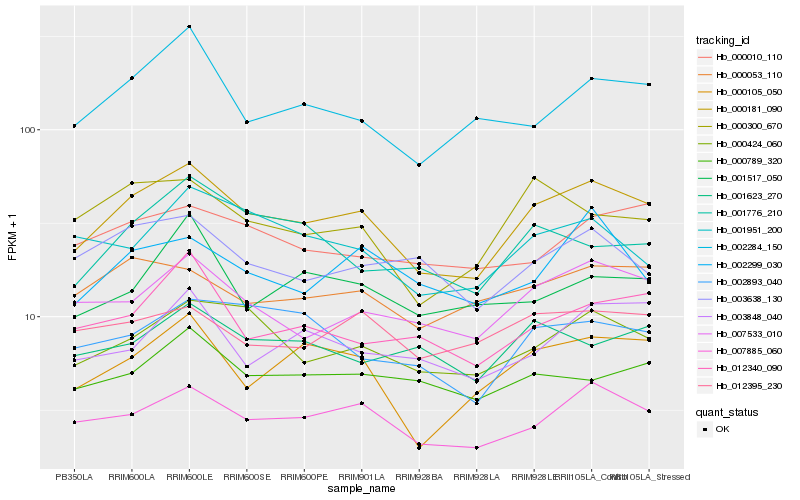
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_090 |
0.0 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 2 |
Hb_007885_060 |
0.0789506862 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 3 |
Hb_000424_060 |
0.0923402 |
- |
- |
PREDICTED: myosin-15 [Jatropha curcas] |
| 4 |
Hb_007533_010 |
0.0953377573 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 5 |
Hb_000105_050 |
0.1015135791 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 6 |
Hb_002284_150 |
0.1093487079 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 7 |
Hb_000053_110 |
0.1106093705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000300_670 |
0.111270842 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001951_200 |
0.1129380269 |
- |
- |
PREDICTED: cold-regulated 413 inner membrane protein 1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000789_320 |
0.1130167152 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_012340_090 |
0.1131415185 |
- |
- |
PREDICTED: endonuclease III homolog 1, chloroplastic isoform X1 [Populus euphratica] |
| 12 |
Hb_012395_230 |
0.113961698 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 13 |
Hb_002893_040 |
0.1142425094 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003638_130 |
0.1142708137 |
- |
- |
PREDICTED: uncharacterized protein LOC105645779 [Jatropha curcas] |
| 15 |
Hb_000010_110 |
0.1147795695 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002299_030 |
0.1163284072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003848_040 |
0.1177713432 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_001776_210 |
0.1196514545 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 19 |
Hb_001623_270 |
0.1215658866 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 20 |
Hb_001517_050 |
0.1231593893 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |