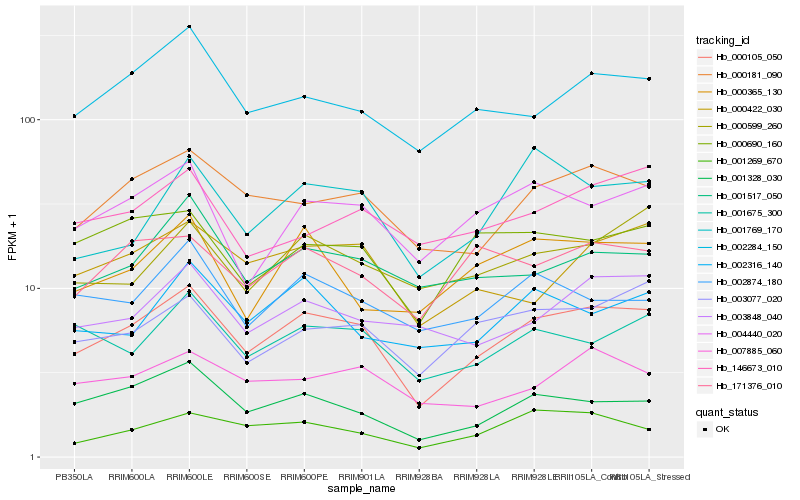
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000105_050 |
0.0 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 2 |
Hb_000181_090 |
0.1015135791 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 3 |
Hb_001328_030 |
0.1079409998 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 1 [Jatropha curcas] |
| 4 |
Hb_004440_020 |
0.1129836425 |
- |
- |
AP-3 complex subunit sigma-1, putative [Ricinus communis] |
| 5 |
Hb_002284_150 |
0.1173204047 |
- |
- |
PREDICTED: uncharacterized protein LOC105635198 [Jatropha curcas] |
| 6 |
Hb_003077_020 |
0.1176589579 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000422_030 |
0.1196468907 |
- |
- |
PREDICTED: uncharacterized protein LOC105643943 [Jatropha curcas] |
| 8 |
Hb_001675_300 |
0.1200976409 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000690_160 |
0.123861361 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 10 |
Hb_001269_670 |
0.124222444 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 11 |
Hb_007885_060 |
0.1242246296 |
- |
- |
replication factor C 40 kDa family protein [Populus trichocarpa] |
| 12 |
Hb_001517_050 |
0.1254962802 |
transcription factor |
TF Family: BBR-BPC |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_171376_010 |
0.1259423497 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003848_040 |
0.1268096207 |
- |
- |
PREDICTED: phosphatidylinositol-glycan biosynthesis class X protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_002874_180 |
0.1279781142 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001769_170 |
0.1292877734 |
- |
- |
50S ribosomal protein L31, putative [Ricinus communis] |
| 17 |
Hb_000365_130 |
0.129899094 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 18 |
Hb_002316_140 |
0.1299167269 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 19 |
Hb_000599_260 |
0.1301225913 |
- |
- |
Vacuolar protein sorting-associated protein 2 like 3 [Glycine soja] |
| 20 |
Hb_146673_010 |
0.1330688554 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |