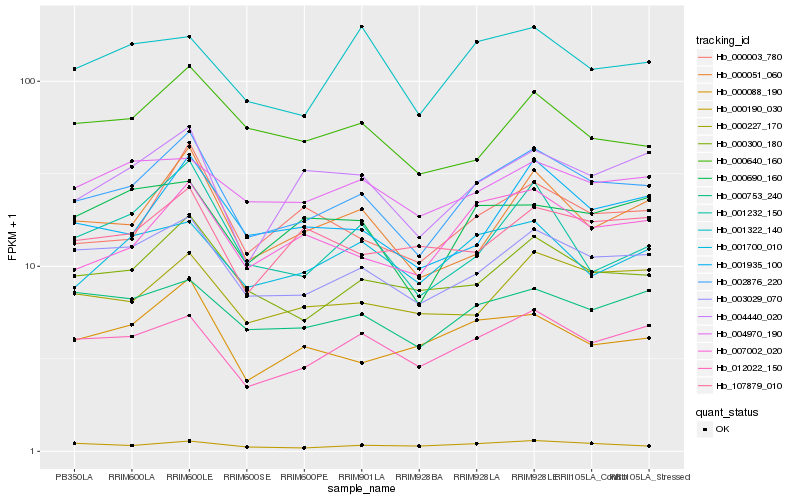
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002876_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 2 |
Hb_003029_070 |
0.0681190891 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000300_180 |
0.0851480785 |
- |
- |
transcription initiation factor brf1, putative [Ricinus communis] |
| 4 |
Hb_012022_150 |
0.0853227829 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 5 |
Hb_007002_020 |
0.0905730302 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 6 |
Hb_000051_060 |
0.0914610298 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001935_100 |
0.0936506957 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 8 |
Hb_004440_020 |
0.093721991 |
- |
- |
AP-3 complex subunit sigma-1, putative [Ricinus communis] |
| 9 |
Hb_000003_780 |
0.0970819064 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_000753_240 |
0.0982901395 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 11 |
Hb_000227_170 |
0.0992416071 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 12 |
Hb_001700_010 |
0.1041960578 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 13 |
Hb_000088_190 |
0.1046721107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000690_160 |
0.1051522812 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 15 |
Hb_000190_030 |
0.1053917786 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsA-like [Pyrus x bretschneideri] |
| 16 |
Hb_004970_190 |
0.1091435056 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000640_160 |
0.1103990415 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 18 |
Hb_001232_150 |
0.1105373431 |
- |
- |
PREDICTED: MAR-binding filament-like protein 1-1 [Jatropha curcas] |
| 19 |
Hb_107879_010 |
0.112736719 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 20 |
Hb_001322_140 |
0.1127512325 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |