| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
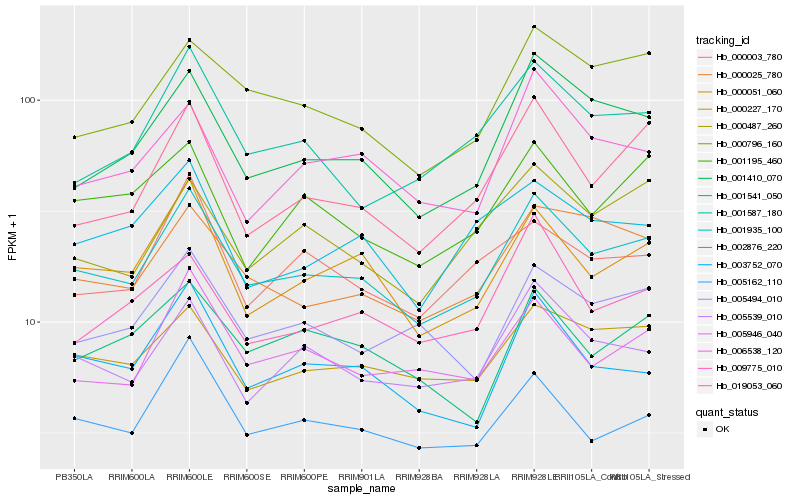
Hb_001935_100 |
0.0 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 2 |
Hb_000051_060 |
0.067841861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000227_170 |
0.0733544741 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_005539_010 |
0.0746839081 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 5 |
Hb_001410_070 |
0.0767013137 |
- |
- |
hypothetical protein JCGZ_20421 [Jatropha curcas] |
| 6 |
Hb_000796_160 |
0.0789987576 |
- |
- |
PREDICTED: nifU-like protein 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005946_040 |
0.0804254427 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000025_780 |
0.0810218802 |
- |
- |
PREDICTED: sanguinarine reductase [Jatropha curcas] |
| 9 |
Hb_005162_110 |
0.0823410834 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 10 |
Hb_003752_070 |
0.0827581141 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 11 |
Hb_019053_060 |
0.0844904975 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 12 |
Hb_005494_010 |
0.0870294832 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_006538_120 |
0.0878488602 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001195_460 |
0.0887982541 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000487_260 |
0.0896685498 |
- |
- |
PREDICTED: protein Iojap, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001587_180 |
0.0910128578 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 17 |
Hb_000003_780 |
0.0924596578 |
- |
- |
hexokinase [Manihot esculenta] |
| 18 |
Hb_009775_010 |
0.0930529976 |
- |
- |
PREDICTED: uncharacterized protein LOC105633555 [Jatropha curcas] |
| 19 |
Hb_002876_220 |
0.0936506957 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 20 |
Hb_001541_050 |
0.0936977258 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |