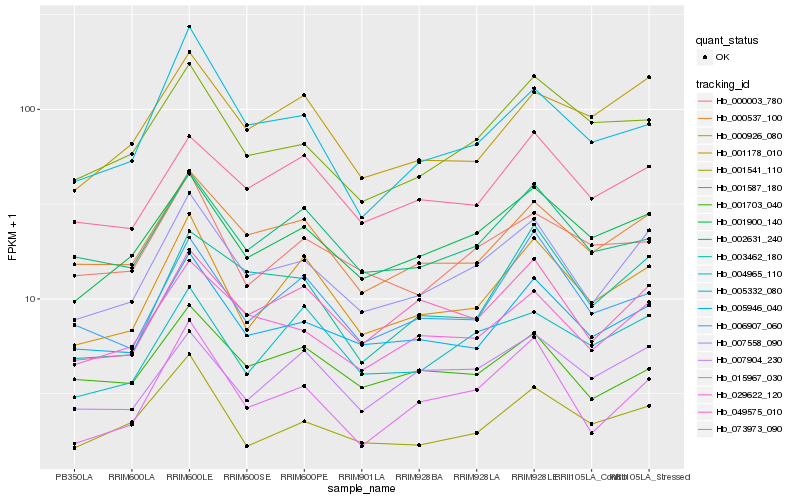
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007558_090 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_029622_120 |
0.0668274472 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 3 |
Hb_001900_140 |
0.0731980245 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005946_040 |
0.0782412813 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000537_100 |
0.079812741 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_049575_010 |
0.0810555521 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 7 |
Hb_000926_080 |
0.0860900985 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 8 |
Hb_015967_030 |
0.0977581501 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001587_180 |
0.0988668456 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 10 |
Hb_073973_090 |
0.0993972674 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 11 |
Hb_002631_240 |
0.1006126949 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 12 |
Hb_004965_110 |
0.1021877333 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 13 |
Hb_007904_230 |
0.1028608425 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001178_010 |
0.1029266952 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 15 |
Hb_001703_040 |
0.1054318501 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003462_180 |
0.1055943753 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 17 |
Hb_006907_060 |
0.107494649 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000003_780 |
0.1077168224 |
- |
- |
hexokinase [Manihot esculenta] |
| 19 |
Hb_001541_110 |
0.1078003005 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_005332_080 |
0.1084449652 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |