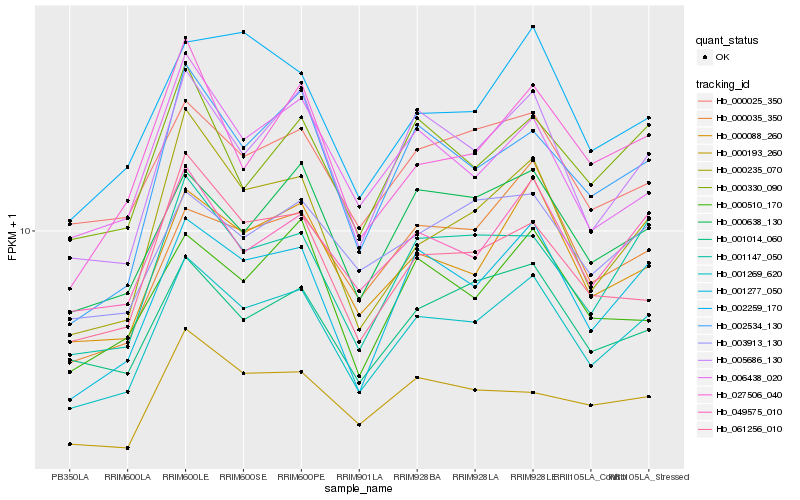
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_620 |
0.0 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 2 |
Hb_001277_050 |
0.0601591736 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005686_130 |
0.0694731256 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 4 |
Hb_000088_260 |
0.0697943124 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 5 |
Hb_049575_010 |
0.078301197 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 6 |
Hb_001147_050 |
0.0797369399 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000235_070 |
0.08338495 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 8 |
Hb_000638_130 |
0.0857827849 |
- |
- |
PREDICTED: SAL1 phosphatase-like [Jatropha curcas] |
| 9 |
Hb_002534_130 |
0.0889897692 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 10 |
Hb_001014_060 |
0.0922059955 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 11 |
Hb_000025_350 |
0.0937754499 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000330_090 |
0.0964222979 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000035_350 |
0.0968091483 |
- |
- |
PREDICTED: monogalactosyldiacylglycerol synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_061256_010 |
0.0986627762 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 15 |
Hb_027506_040 |
0.0988130909 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003913_130 |
0.0991128666 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 17 |
Hb_002259_170 |
0.1013227332 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000510_170 |
0.1019546447 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 19 |
Hb_006438_020 |
0.1039935719 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 20 |
Hb_000193_260 |
0.1044230829 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |