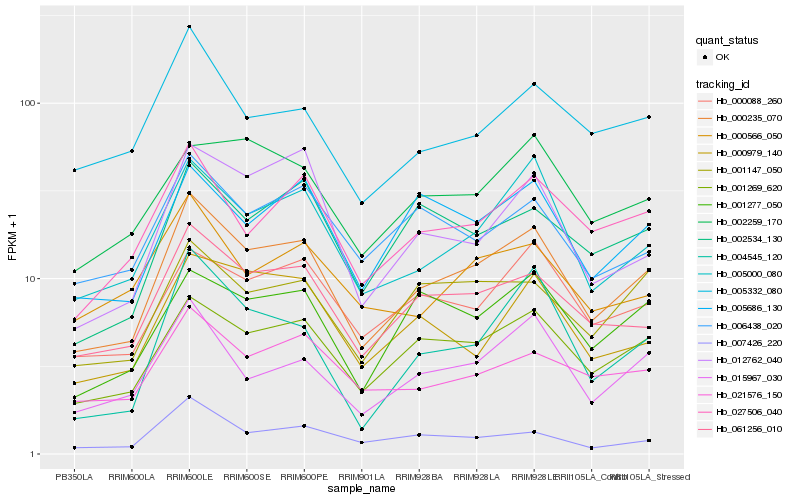
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000235_070 |
0.0 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 2 |
Hb_001269_620 |
0.08338495 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 3 |
Hb_061256_010 |
0.0894198935 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005000_080 |
0.0976487818 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 5 |
Hb_027506_040 |
0.1068986544 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_015967_030 |
0.1075535113 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000088_260 |
0.1146248227 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 8 |
Hb_021576_150 |
0.1175915768 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 9 |
Hb_001147_050 |
0.1181518884 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005686_130 |
0.1183014827 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 11 |
Hb_001277_050 |
0.1192529065 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005332_080 |
0.1198242427 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 13 |
Hb_002534_130 |
0.1213168983 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 14 |
Hb_006438_020 |
0.1250076059 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 15 |
Hb_000979_140 |
0.1263796406 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 16 |
Hb_012762_040 |
0.1276827264 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 17 |
Hb_004545_120 |
0.1281140662 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 18 |
Hb_000566_050 |
0.129721879 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 19 |
Hb_007426_220 |
0.1301750663 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_002259_170 |
0.1306845622 |
- |
- |
PREDICTED: internal alternative NAD(P)H-ubiquinone oxidoreductase A1, mitochondrial-like [Jatropha curcas] |