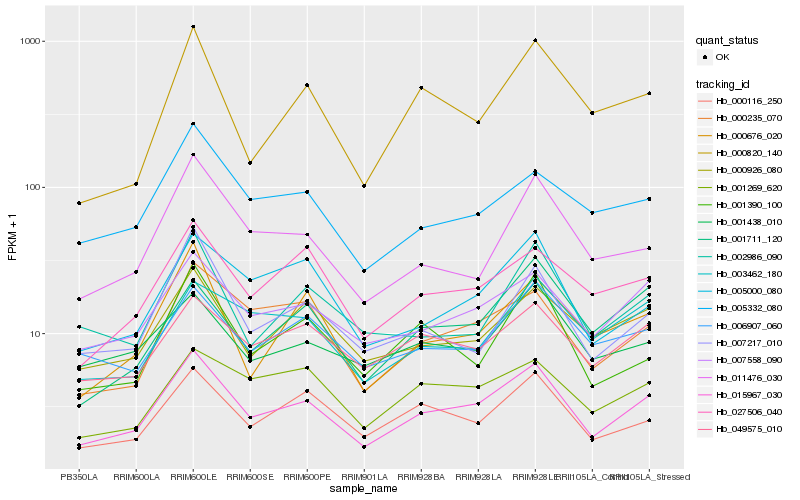
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015967_030 |
0.0 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007558_090 |
0.0977581501 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_000235_070 |
0.1075535113 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 4 |
Hb_000116_250 |
0.1102411054 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 5 |
Hb_000926_080 |
0.114561437 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 6 |
Hb_027506_040 |
0.1151829002 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_049575_010 |
0.1167716062 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 8 |
Hb_005000_080 |
0.1187404177 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 9 |
Hb_005332_080 |
0.1190412045 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_007217_010 |
0.1197314305 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000676_020 |
0.12092113 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 12 |
Hb_011476_030 |
0.1235130004 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 13 |
Hb_001269_620 |
0.1241960514 |
- |
- |
PREDICTED: magnesium transporter MRS2-1 isoform X1 [Populus euphratica] |
| 14 |
Hb_002986_090 |
0.1261991456 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_001390_100 |
0.1275672592 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_006907_060 |
0.1276087816 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000820_140 |
0.1280589137 |
- |
- |
histone H4 [Zea mays] |
| 18 |
Hb_001438_010 |
0.128682505 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003462_180 |
0.128999635 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 20 |
Hb_001711_120 |
0.1294029894 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |