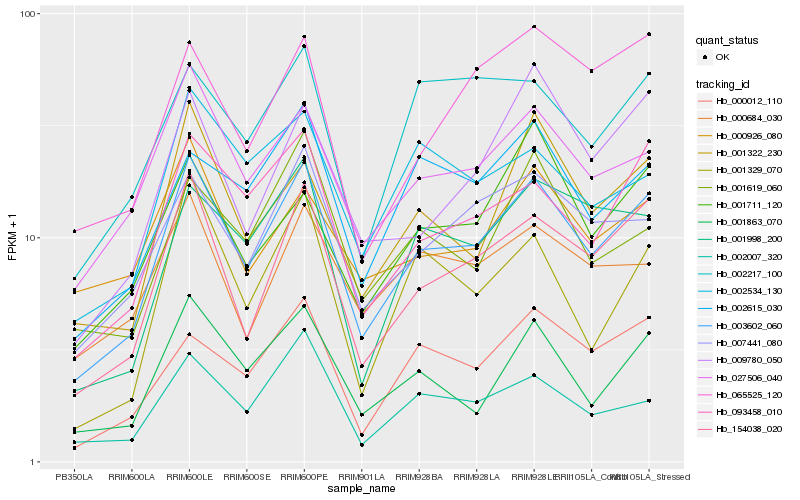
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003602_060 |
0.0 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 2 |
Hb_154038_020 |
0.0804204448 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 3 |
Hb_093458_010 |
0.0977527334 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 4 |
Hb_027506_040 |
0.0978693023 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_007441_080 |
0.1060107625 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 6 |
Hb_001863_070 |
0.107494376 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 7 |
Hb_002007_320 |
0.1135730245 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 8 |
Hb_001322_230 |
0.1152950457 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 9 |
Hb_000012_110 |
0.1153245197 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 10 |
Hb_001998_200 |
0.1193352348 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 11 |
Hb_002615_030 |
0.1207932405 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 12 |
Hb_009780_050 |
0.1255272433 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_065525_120 |
0.1258995751 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 14 |
Hb_001619_060 |
0.1266322764 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000684_030 |
0.1267596647 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_002217_100 |
0.1275219965 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_001711_120 |
0.1279086883 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001329_070 |
0.1282099164 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 19 |
Hb_000926_080 |
0.1286062856 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 20 |
Hb_002534_130 |
0.1292741586 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |