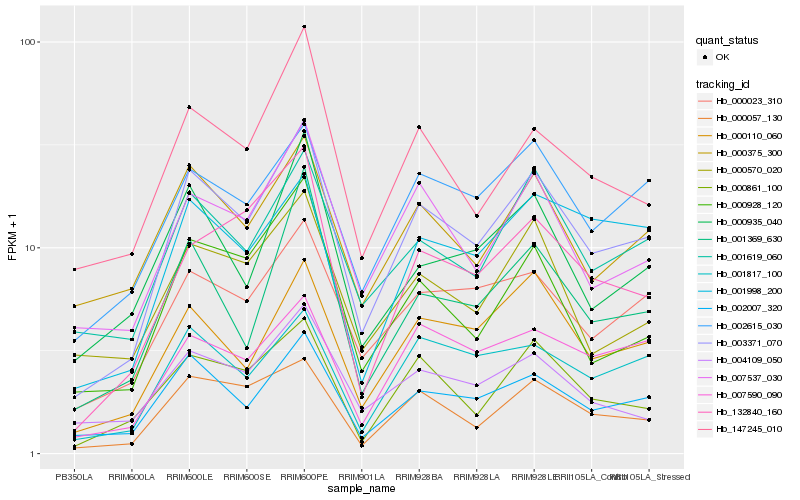
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003371_070 |
0.0 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_002007_320 |
0.0902307105 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 3 |
Hb_000057_130 |
0.102267965 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 4 |
Hb_000110_060 |
0.1026911135 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 5 |
Hb_001619_060 |
0.1073650361 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000023_310 |
0.1088346792 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |
| 7 |
Hb_007537_030 |
0.109123103 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000861_100 |
0.1129690257 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002615_030 |
0.1160081886 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 10 |
Hb_001817_100 |
0.1179283926 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 11 |
Hb_000375_300 |
0.1194749795 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 12 |
Hb_000928_120 |
0.120204722 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 13 |
Hb_147245_010 |
0.1204622474 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 14 |
Hb_001369_630 |
0.1214976918 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 15 |
Hb_007590_090 |
0.1243003177 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 16 |
Hb_000935_040 |
0.1260155198 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_132840_160 |
0.1282259341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000570_020 |
0.1285713917 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 19 |
Hb_001998_200 |
0.1293953369 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 20 |
Hb_004109_050 |
0.1317379383 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |