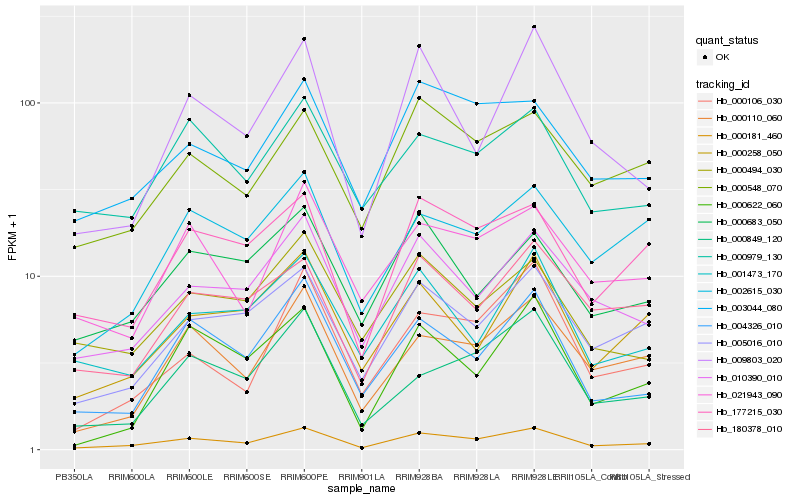
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_460 |
0.0 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 2 |
Hb_004326_010 |
0.1077939147 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_005016_010 |
0.1080645311 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_010390_010 |
0.1095764886 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 5 |
Hb_000110_060 |
0.1119362402 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 6 |
Hb_177215_030 |
0.1158151979 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 7 |
Hb_003044_080 |
0.1230381554 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_000548_070 |
0.1233696594 |
desease resistance |
Gene Name: Synthase_beta |
RecName: Full=ATP synthase subunit beta, mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 9 |
Hb_000106_030 |
0.1254847365 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000258_050 |
0.1256877576 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_009803_020 |
0.1271600662 |
- |
- |
hypothetical protein PRUPE_ppa006317mg [Prunus persica] |
| 12 |
Hb_180378_010 |
0.127620364 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 13 |
Hb_000849_120 |
0.1282130905 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 14 |
Hb_001473_170 |
0.1282354675 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 15 |
Hb_000979_130 |
0.1294595784 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_002615_030 |
0.1300024213 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 17 |
Hb_021943_090 |
0.1304583465 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 18 |
Hb_000622_060 |
0.1328420284 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000683_050 |
0.1344196419 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 20 |
Hb_000494_030 |
0.1344834577 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |