| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
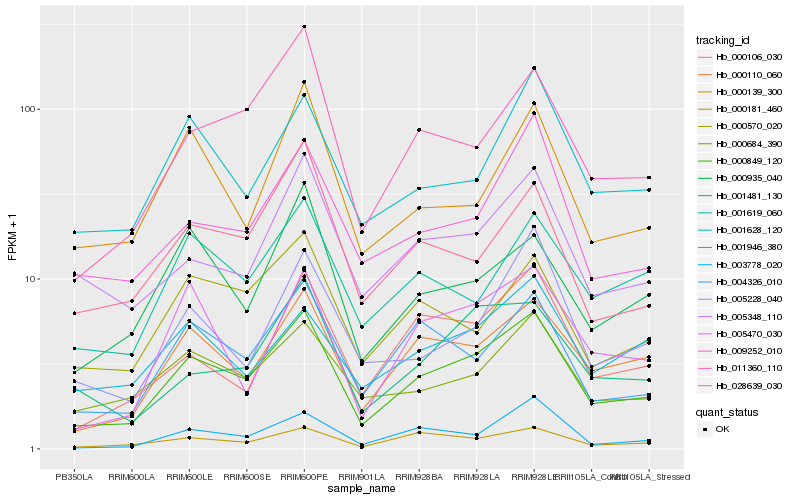
Hb_000849_120 |
0.0 |
- |
- |
PREDICTED: linoleate 9S-lipoxygenase 6-like [Jatropha curcas] |
| 2 |
Hb_000110_060 |
0.1104048081 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 3 |
Hb_000684_390 |
0.1271946161 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 4 |
Hb_004326_010 |
0.1274319767 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_000181_460 |
0.1282130905 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 6 |
Hb_009252_010 |
0.1368188338 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 7 |
Hb_005348_110 |
0.1371183552 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_011360_110 |
0.1377361575 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1 [Vitis vinifera] |
| 9 |
Hb_001628_120 |
0.1391887985 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 10 |
Hb_000106_030 |
0.1392444646 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_000570_020 |
0.1406510361 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 12 |
Hb_001481_130 |
0.1426536848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003778_020 |
0.1433910208 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_005470_030 |
0.1434115024 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005228_040 |
0.1459927638 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 16 |
Hb_028639_030 |
0.1474605278 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 17 |
Hb_001619_060 |
0.1474831313 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000139_300 |
0.1496834588 |
- |
- |
hypothetical protein POPTR_0008s19950g [Populus trichocarpa] |
| 19 |
Hb_001946_380 |
0.1506106384 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 20 |
Hb_000935_040 |
0.1510455359 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |