| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
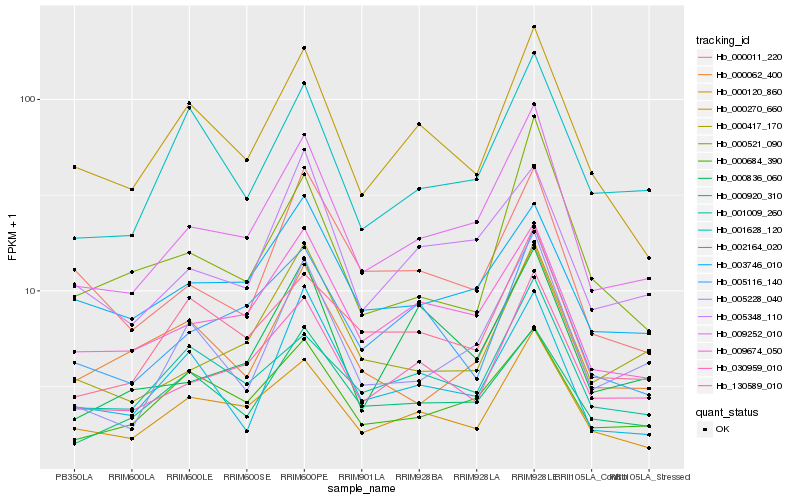
Hb_009252_010 |
0.0 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 2 |
Hb_030959_010 |
0.0901115025 |
- |
- |
cullin, putative [Ricinus communis] |
| 3 |
Hb_009674_050 |
0.0928322851 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 4 |
Hb_000120_860 |
0.1001947768 |
- |
- |
nucellin, putative [Ricinus communis] |
| 5 |
Hb_005228_040 |
0.1013067166 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 6 |
Hb_000684_390 |
0.1119495344 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 7 |
Hb_005348_110 |
0.1119990067 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_000417_170 |
0.1139252855 |
- |
- |
PREDICTED: uncharacterized protein LOC105649689 [Jatropha curcas] |
| 9 |
Hb_130589_010 |
0.1185134074 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_000270_660 |
0.1207882584 |
- |
- |
purine permease, putative [Ricinus communis] |
| 11 |
Hb_003746_010 |
0.1232866447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001009_260 |
0.1254474658 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001628_120 |
0.1254489182 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 14 |
Hb_000920_310 |
0.1271811062 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_000062_400 |
0.128554987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005116_140 |
0.1300113634 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 17 |
Hb_000011_220 |
0.1321658247 |
- |
- |
PREDICTED: uncharacterized protein LOC105631313 [Jatropha curcas] |
| 18 |
Hb_000836_060 |
0.1328730854 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 19 |
Hb_000521_090 |
0.1339101984 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002164_020 |
0.1343585146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |