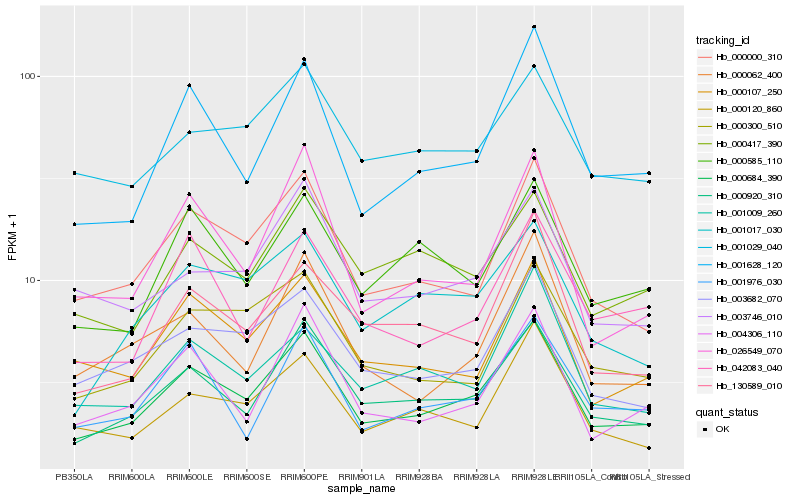
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_390 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 2 |
Hb_000920_310 |
0.0672467117 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 3 |
Hb_000000_310 |
0.0791818362 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 4 |
Hb_001628_120 |
0.0847097709 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 5 |
Hb_003682_070 |
0.0914118266 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000107_250 |
0.0986834781 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 7 |
Hb_001017_030 |
0.0999150112 |
- |
- |
copine, putative [Ricinus communis] |
| 8 |
Hb_004306_110 |
0.1021223185 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 9 |
Hb_026549_070 |
0.102239046 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 10 |
Hb_003746_010 |
0.1022726514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000300_510 |
0.1024165207 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 12 |
Hb_000120_860 |
0.1029687112 |
- |
- |
nucellin, putative [Ricinus communis] |
| 13 |
Hb_130589_010 |
0.1034065149 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 14 |
Hb_000585_110 |
0.1083950353 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 15 |
Hb_000062_400 |
0.1091875169 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001976_030 |
0.1093094141 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001029_040 |
0.1095566651 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 18 |
Hb_000417_390 |
0.1095853542 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 19 |
Hb_001009_260 |
0.1115606343 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_042083_040 |
0.1117746127 |
- |
- |
conserved hypothetical protein [Ricinus communis] |