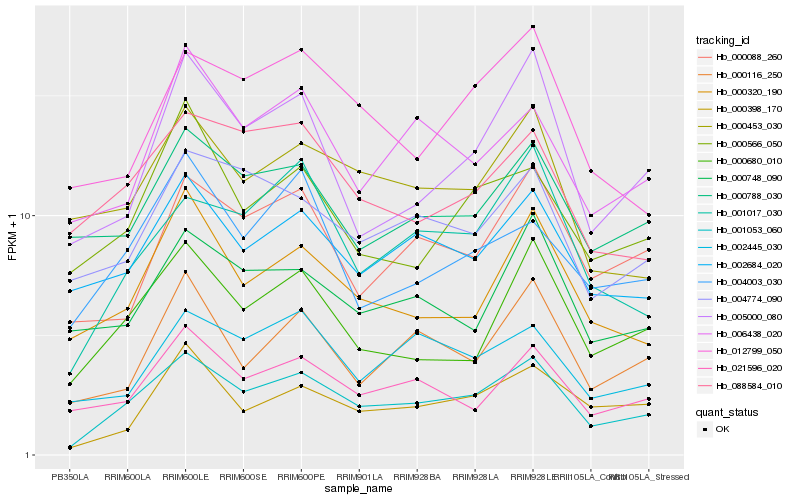
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001053_060 |
0.0 |
- |
- |
DHHC-type zinc finger family protein [Theobroma cacao] |
| 2 |
Hb_001017_030 |
0.1174122138 |
- |
- |
copine, putative [Ricinus communis] |
| 3 |
Hb_000680_010 |
0.1241060206 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 4 |
Hb_002684_020 |
0.1265148705 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 5 |
Hb_000398_170 |
0.1275113994 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1-like [Jatropha curcas] |
| 6 |
Hb_000320_190 |
0.1299595072 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_021596_020 |
0.130763638 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 8 |
Hb_004774_090 |
0.1320784175 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Jatropha curcas] |
| 9 |
Hb_005000_080 |
0.1328368626 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 10 |
Hb_000088_260 |
0.1330185848 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 11 |
Hb_000116_250 |
0.1347706678 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 12 |
Hb_006438_020 |
0.1364706595 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 13 |
Hb_002445_030 |
0.137158715 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 14 |
Hb_088584_010 |
0.1380742299 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 15 |
Hb_012799_050 |
0.138837473 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 16 |
Hb_000788_030 |
0.1388894961 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 17 |
Hb_004003_030 |
0.1390218809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000453_030 |
0.1395597778 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 19 |
Hb_000748_090 |
0.1396152379 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 20 |
Hb_000566_050 |
0.1401829422 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |