| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
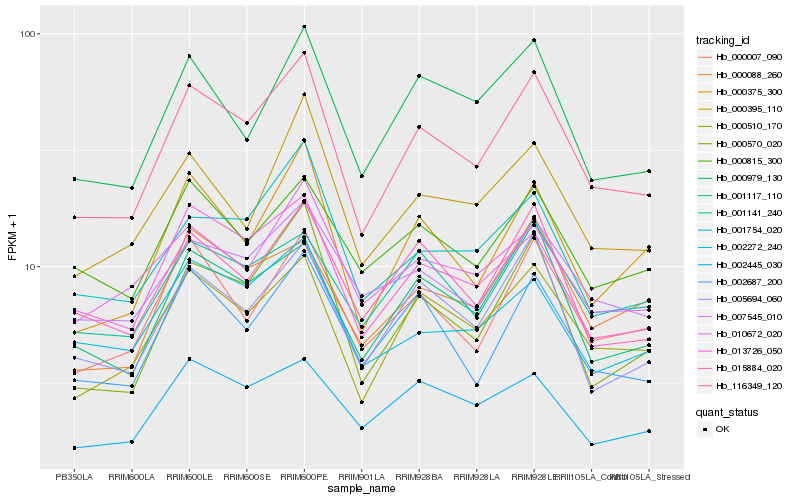
Hb_116349_120 |
0.0 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 2 |
Hb_000510_170 |
0.058888892 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 3 |
Hb_001117_110 |
0.0741477371 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 4 |
Hb_013726_050 |
0.0754334169 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 5 |
Hb_005694_060 |
0.079725521 |
- |
- |
PREDICTED: bifunctional fucokinase/fucose pyrophosphorylase [Jatropha curcas] |
| 6 |
Hb_000570_020 |
0.0802590244 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 7 |
Hb_000375_300 |
0.0820838999 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 8 |
Hb_000007_090 |
0.0821199486 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 9 |
Hb_015884_020 |
0.0828190596 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 10 |
Hb_002687_200 |
0.0841952719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000979_130 |
0.0843632191 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 12 |
Hb_000395_110 |
0.0865076542 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 13 |
Hb_001754_020 |
0.0865949716 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |
| 14 |
Hb_007545_010 |
0.0885938238 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 15 |
Hb_000088_260 |
0.0897423637 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 16 |
Hb_001141_240 |
0.0923163623 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002445_030 |
0.0923294559 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 18 |
Hb_010672_020 |
0.0937491572 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000815_300 |
0.094759383 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 20 |
Hb_002272_240 |
0.0951248373 |
- |
- |
catalytic, putative [Ricinus communis] |