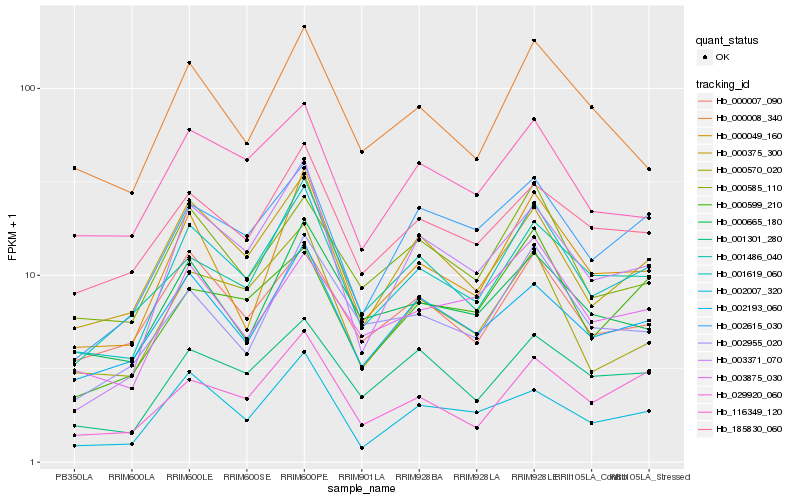
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001619_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000375_300 |
0.080642852 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 3 |
Hb_000049_160 |
0.0880694231 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 4 |
Hb_029920_060 |
0.0933289214 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001301_280 |
0.0962294284 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002193_060 |
0.0979467724 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 7 |
Hb_185830_060 |
0.0993197922 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 8 |
Hb_000585_110 |
0.0995889923 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 9 |
Hb_000007_090 |
0.1005707706 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 10 |
Hb_000570_020 |
0.1006588416 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 11 |
Hb_002615_030 |
0.1028736003 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 12 |
Hb_116349_120 |
0.1047554832 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit alpha-1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_002955_020 |
0.1059769238 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_002007_320 |
0.1063472296 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 15 |
Hb_001486_040 |
0.1064154492 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003371_070 |
0.1073650361 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000665_180 |
0.1078631381 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 18 |
Hb_000008_340 |
0.10902171 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 19 |
Hb_003875_030 |
0.1092054759 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 20 |
Hb_000599_210 |
0.1105457836 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |