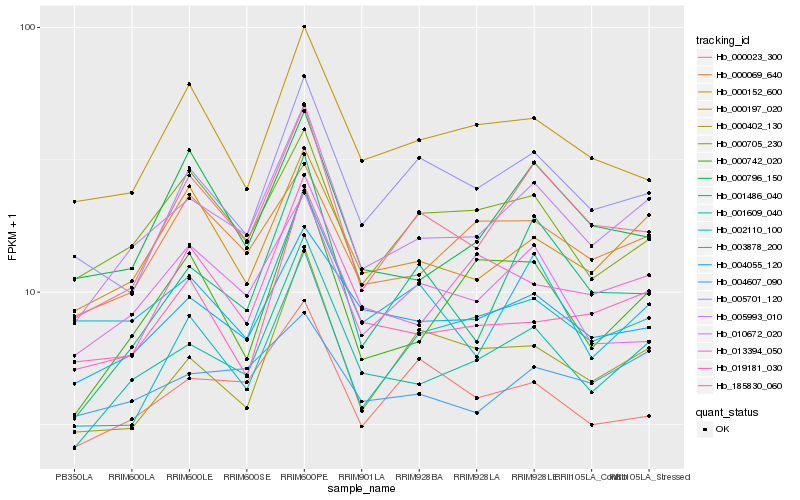
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005993_010 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 2 |
Hb_001609_040 |
0.0512057588 |
- |
- |
hypothetical protein POPTR_0017s05650g [Populus trichocarpa] |
| 3 |
Hb_185830_060 |
0.0802021786 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 4 |
Hb_000152_600 |
0.0860880105 |
- |
- |
PREDICTED: protein jagunal homolog 1 [Jatropha curcas] |
| 5 |
Hb_004055_120 |
0.0888136657 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 6 |
Hb_000069_640 |
0.0950005528 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 7 |
Hb_000705_230 |
0.098750088 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 8 |
Hb_013394_050 |
0.1014932631 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 9 |
Hb_004607_090 |
0.1033201932 |
- |
- |
PREDICTED: uncharacterized protein LOC105648257 [Jatropha curcas] |
| 10 |
Hb_000742_020 |
0.1034337739 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 11 |
Hb_005701_120 |
0.1046010454 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 12 |
Hb_010672_020 |
0.1051219989 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000402_130 |
0.1052403229 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 14 |
Hb_000796_150 |
0.105737901 |
- |
- |
PREDICTED: uncharacterized protein LOC105136996 isoform X1 [Populus euphratica] |
| 15 |
Hb_001486_040 |
0.1061241923 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 16 |
Hb_019181_030 |
0.1066968681 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 17 |
Hb_000023_300 |
0.1070624319 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 18 |
Hb_002110_100 |
0.1090492259 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000197_020 |
0.109943112 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 20 |
Hb_003878_200 |
0.1105139512 |
- |
- |
PREDICTED: V-type proton ATPase subunit D-like [Jatropha curcas] |