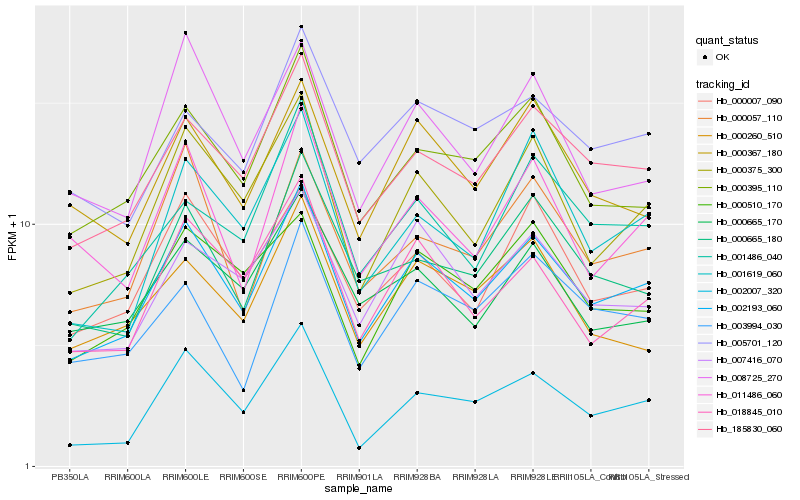
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002193_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 2 |
Hb_185830_060 |
0.0604174068 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 3 |
Hb_000375_300 |
0.0694840683 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 4 |
Hb_000395_110 |
0.0800165083 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 5 |
Hb_018845_010 |
0.0858814035 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 6 |
Hb_000007_090 |
0.0864940644 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 7 |
Hb_007416_070 |
0.0876388071 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 8 |
Hb_002007_320 |
0.0891677644 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase ANP1 [Jatropha curcas] |
| 9 |
Hb_000665_180 |
0.0907570445 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 10 |
Hb_000057_110 |
0.092501176 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 11 |
Hb_000367_180 |
0.0950338427 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 12 |
Hb_000510_170 |
0.0955729176 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 13 |
Hb_005701_120 |
0.0966388916 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 14 |
Hb_011486_060 |
0.0977434394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001619_060 |
0.0979467724 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001486_040 |
0.0980736172 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008725_270 |
0.0991212504 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 18 |
Hb_000260_510 |
0.0992764091 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000665_170 |
0.0996821528 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 20 |
Hb_003994_030 |
0.100857803 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |