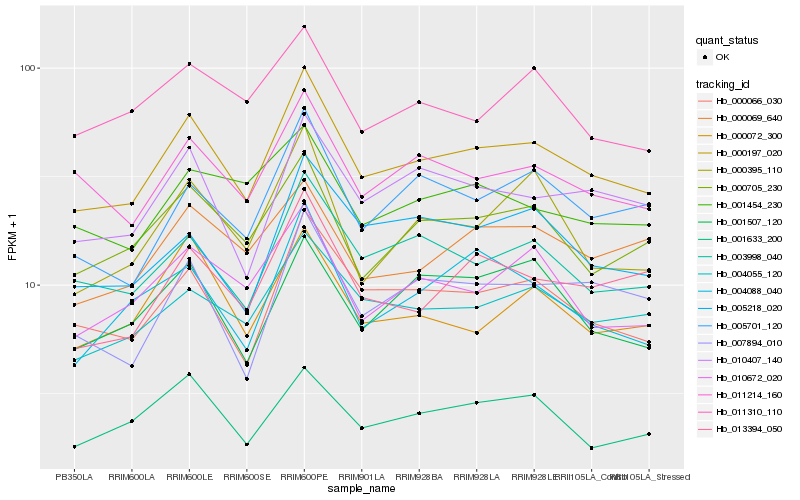
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000197_020 |
0.0 |
- |
- |
PREDICTED: protein RER1A [Jatropha curcas] |
| 2 |
Hb_000066_030 |
0.0656518883 |
- |
- |
PREDICTED: uncharacterized protein LOC105643898 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003998_040 |
0.0739731167 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 4 |
Hb_004055_120 |
0.074668193 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 5 |
Hb_000705_230 |
0.0791921958 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 6 |
Hb_010407_140 |
0.0798534603 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 7 |
Hb_011214_160 |
0.079861614 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 8 |
Hb_013394_050 |
0.0854445615 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2 [Nicotiana sylvestris] |
| 9 |
Hb_010672_020 |
0.0859647596 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_005701_120 |
0.0867339408 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 11 |
Hb_007894_010 |
0.089223174 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 12 |
Hb_001633_200 |
0.089252978 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 13 |
Hb_000072_300 |
0.0909996043 |
- |
- |
PREDICTED: probable galacturonosyltransferase 9 [Jatropha curcas] |
| 14 |
Hb_000069_640 |
0.091903033 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 15 |
Hb_005218_020 |
0.0938094383 |
- |
- |
Uncharacterized protein isoform 3 [Theobroma cacao] |
| 16 |
Hb_004088_040 |
0.0944974375 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 17 |
Hb_001507_120 |
0.0947015699 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000395_110 |
0.0954322153 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 19 |
Hb_011310_110 |
0.0964388507 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001454_230 |
0.0973263401 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |