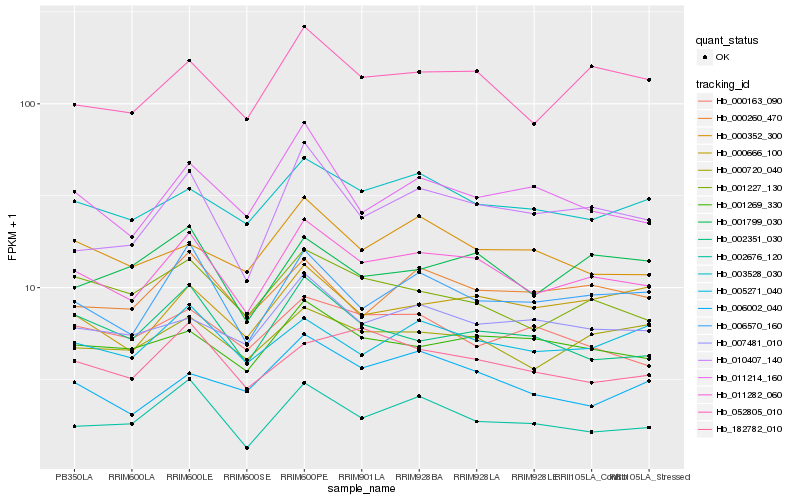
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011282_060 |
0.0 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 2 |
Hb_052805_010 |
0.0724684702 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 3 |
Hb_006570_160 |
0.076244304 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 4 |
Hb_003528_030 |
0.083293509 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 5 |
Hb_001227_130 |
0.0841876702 |
- |
- |
PREDICTED: delta(14)-sterol reductase [Jatropha curcas] |
| 6 |
Hb_001269_330 |
0.0855170528 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 7 |
Hb_010407_140 |
0.0863469208 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 8 |
Hb_000720_040 |
0.0877368486 |
- |
- |
PREDICTED: replication factor C subunit 2 [Jatropha curcas] |
| 9 |
Hb_007481_010 |
0.0898937605 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 10 |
Hb_000260_470 |
0.0914300028 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 11 |
Hb_002351_030 |
0.0924654557 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_011214_160 |
0.0934167306 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 13 |
Hb_000352_300 |
0.0959381492 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 14 |
Hb_005271_040 |
0.0963111197 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 15 |
Hb_000666_100 |
0.0968159695 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 16 |
Hb_182782_010 |
0.0984867482 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002676_120 |
0.0987369099 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 18 |
Hb_006002_040 |
0.1002665795 |
- |
- |
PREDICTED: uncharacterized protein LOC105638085 [Jatropha curcas] |
| 19 |
Hb_001799_030 |
0.1003798278 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 20 |
Hb_000163_090 |
0.1009102755 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |