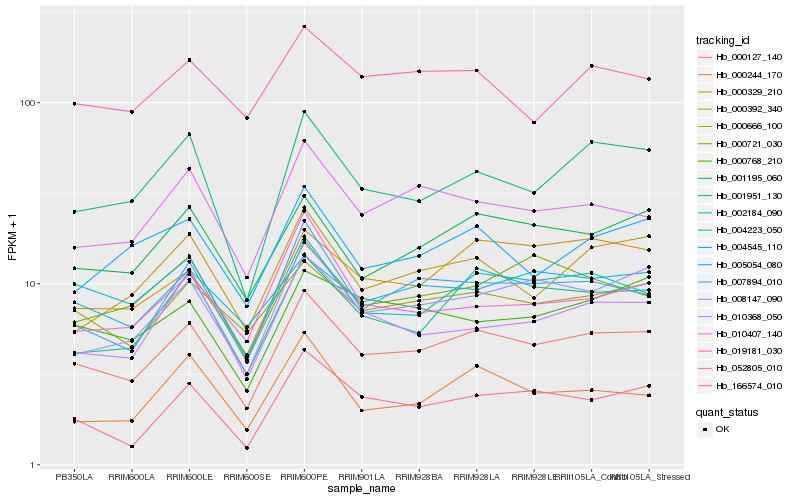
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000127_140 |
0.0 |
- |
- |
transporter-related family protein [Populus trichocarpa] |
| 2 |
Hb_007894_010 |
0.07027183 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 3 |
Hb_008147_090 |
0.077722449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001951_130 |
0.0794627379 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 5 |
Hb_001195_060 |
0.0810324449 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 6 |
Hb_000666_100 |
0.0827397307 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 7 |
Hb_002184_090 |
0.0827513943 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_010407_140 |
0.0842888697 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 9 |
Hb_004223_050 |
0.0856702291 |
- |
- |
hexokinase [Manihot esculenta] |
| 10 |
Hb_010368_050 |
0.0881530464 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 11 |
Hb_000392_340 |
0.0923589069 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 12 |
Hb_166574_010 |
0.0939952609 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |
| 13 |
Hb_000329_210 |
0.0962481861 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_004545_110 |
0.0969497235 |
- |
- |
DHHC-type zinc finger family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000768_210 |
0.09851007 |
- |
- |
PREDICTED: uncharacterized protein LOC105632522 [Jatropha curcas] |
| 16 |
Hb_005054_080 |
0.0988876636 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 17 |
Hb_000721_030 |
0.1004091159 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 18 |
Hb_052805_010 |
0.1029130639 |
- |
- |
hypothetical protein POPTR_0009s03650g [Populus trichocarpa] |
| 19 |
Hb_000244_170 |
0.1053737672 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 20 |
Hb_019181_030 |
0.1054735198 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |