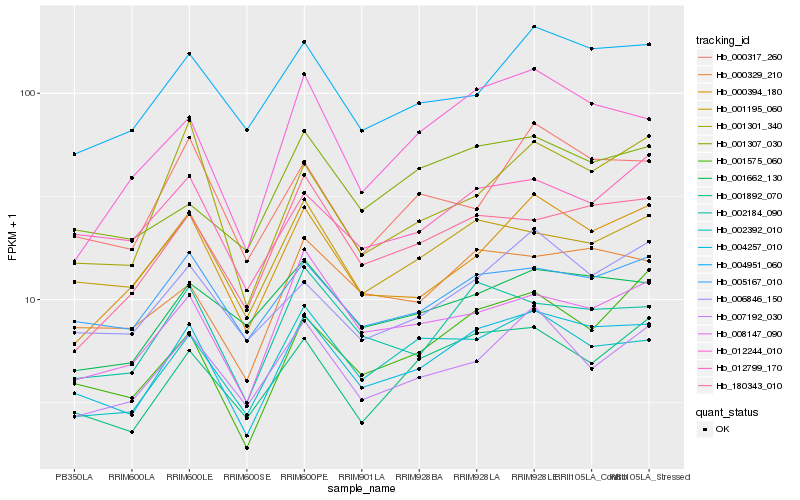
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004257_010 |
0.0 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000329_210 |
0.0855432758 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 3 |
Hb_002392_010 |
0.0885161648 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 4 |
Hb_002184_090 |
0.0886034985 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_001195_060 |
0.0896240479 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 6 |
Hb_005167_010 |
0.0903834148 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001892_070 |
0.0963249259 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 8 |
Hb_004951_060 |
0.0968246761 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 9 |
Hb_000394_180 |
0.0984825394 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 10 |
Hb_008147_090 |
0.1001387094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007192_030 |
0.1021926577 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 12 |
Hb_012799_170 |
0.1022162231 |
- |
- |
PREDICTED: protein GLC8 isoform X2 [Jatropha curcas] |
| 13 |
Hb_180343_010 |
0.1029342578 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 14 |
Hb_000317_260 |
0.1032379931 |
- |
- |
unknown [Populus trichocarpa] |
| 15 |
Hb_012244_010 |
0.1036720538 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 16 |
Hb_006846_150 |
0.1044761931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001662_130 |
0.1045287805 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001575_060 |
0.1067083768 |
- |
- |
PREDICTED: 2-methoxy-6-polyprenyl-1,4-benzoquinol methylase, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_001301_340 |
0.1068945656 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 20 |
Hb_001307_030 |
0.1071847939 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |