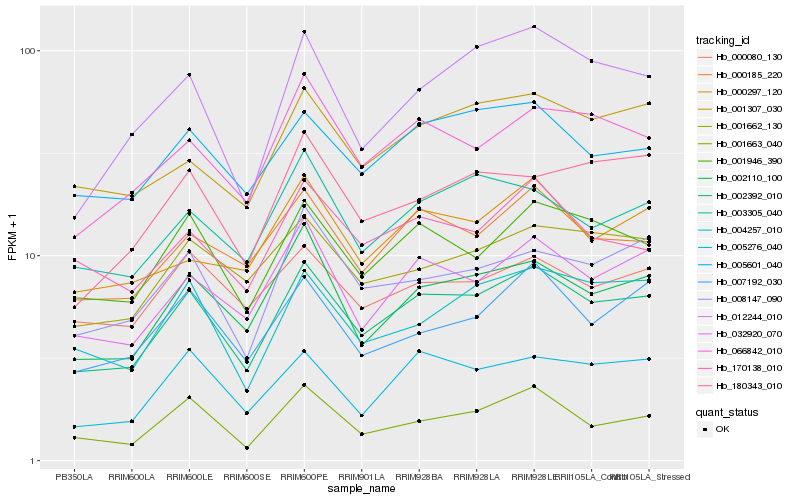
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002392_010 |
0.0 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 2 |
Hb_001946_390 |
0.0697205183 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 3 |
Hb_032920_070 |
0.0758033215 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 4 |
Hb_002110_100 |
0.0760044128 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000185_220 |
0.0778180079 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 6 |
Hb_066842_010 |
0.0799486605 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005601_040 |
0.0822643081 |
- |
- |
Histone deacetylase 1 isoform 1 [Theobroma cacao] |
| 8 |
Hb_005276_040 |
0.0876001064 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 9 |
Hb_001662_130 |
0.0878975029 |
- |
- |
PREDICTED: bifunctional nuclease 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004257_010 |
0.0885161648 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 11 |
Hb_008147_090 |
0.0889043721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001307_030 |
0.0889720619 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 13 |
Hb_003305_040 |
0.0890313584 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 14 |
Hb_012244_010 |
0.0909545276 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 15 |
Hb_170138_010 |
0.0935513329 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007192_030 |
0.0939118076 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 17 |
Hb_000080_130 |
0.0948576751 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 18 |
Hb_000297_120 |
0.095014239 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 19 |
Hb_001663_040 |
0.0954118031 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 20 |
Hb_180343_010 |
0.0963393457 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |