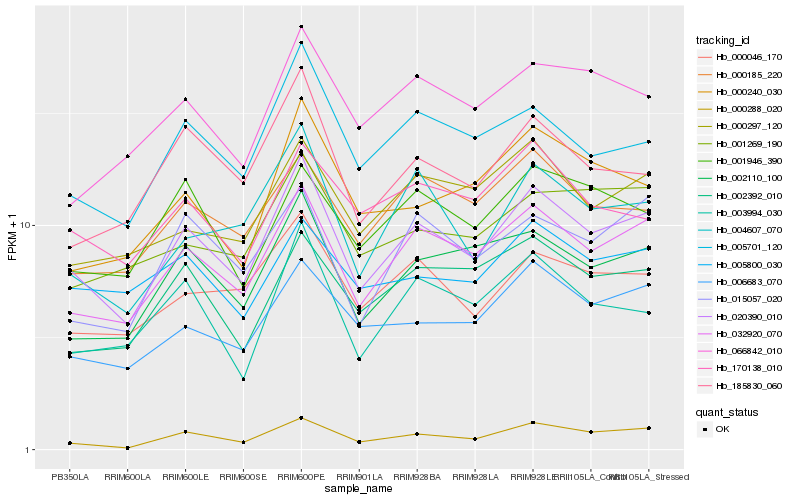
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020390_010 |
0.0 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 2 |
Hb_032920_070 |
0.0587386497 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 3 |
Hb_005701_120 |
0.085599745 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 4 |
Hb_000288_020 |
0.0864463776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002110_100 |
0.0881950569 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_006683_070 |
0.0916420374 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 7 |
Hb_000046_170 |
0.0938412521 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004607_070 |
0.0967465282 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 9 |
Hb_185830_060 |
0.1005861803 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000185_220 |
0.1013155402 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 11 |
Hb_003994_030 |
0.1016781036 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 12 |
Hb_002392_010 |
0.1048093156 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 13 |
Hb_000297_120 |
0.1048657104 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 14 |
Hb_015057_020 |
0.1054795067 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 15 |
Hb_005800_030 |
0.1071375746 |
- |
- |
PREDICTED: lysM and putative peptidoglycan-binding domain-containing protein 4-like [Jatropha curcas] |
| 16 |
Hb_170138_010 |
0.1076218134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_066842_010 |
0.1082285512 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001946_390 |
0.1083879764 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 19 |
Hb_001269_190 |
0.1086070472 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 20 |
Hb_000240_030 |
0.1091534338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |